The objectives of this study were to determine the serovar of a collection of Actinobacillus pleuropneumoniae strains within the 3-6-8-15 cross-reacting group and to analyze their phenotypic and genetic properties. Based on the serological tests, forty-seven field strains of Actinobacillus pleuropneumoniae isolated from lungs with pleuropneumonia lesions in Japan and Argentina were found to be serovars belonging to the 3-6-8-15 cross-reacting group. By using a capsule loci-based PCR, twenty-nine (96.7%) and one (3.3%) from Japan were identified as serovars 15 and 8, respectively, whereas seventeen (100%) from Argentina were identified as serovar 8. The findings suggested that serovars 8 and 15 were prevalent within the 3-6-8-15 cross-reacting group, in Argentina and Japan, respectively. Phenotypic analyses revealed that the protein patterns observed on SDS–PAGE and the lipopolysaccharide antigen detected by immunoblotting of the reference and field strains of serovars 8 and 15 were similar to each other. Genetic (16S rDNA, apxIIA, apxIIIA, cps, cpx genes, apx and omlA patterns) analyses revealed that the apxIIA and apxIIIA genes of the field strains of serovars 8 and 15 were similar to those of the reference strains of serovars 3, 4, 6, 8 and 15. The results obtained in the present study may be useful for the development of more effective vaccines against disease caused by A. pleuropneumoniae by including the homologous antigens to the most prevalent serovars in specific geographical areas.
Los objetivos del presente estudio fueron determinar el serovar de una colección de cepas de Actinobacillus pleuropneumoniae pertenecientes al grupo 3, 6, 8, 15 de reacciones cruzadas y analizar sus propiedades fenotípicas y genéticas. En base a técnicas serológicas se determinó que cuarenta y siete cepas de A. pleuropneumoniae aisladas a partir de pulmones con lesiones de pleuroneumonía en Japón y Argentina pertenecen al grupo 3, 6, 8, 15. Mediante el uso de PCR basado en locus capsulares, veintinueve (96.7%) y una (3.3%) de los aislados japoneses fueron identificados como serovar 15 y 8 respectivamente, mientras que diecisiete (100%) de los aislados argentinos resultaron pertenecer al serotipo 8. Este hallazgo sugirió que los serovares 8 y 15 fueron los prevalentes dentro del grupo 3, 6, 8, 15 en Japón y Argentina, respectivamente. El análisis fenotípico reveló que los perfiles proteicos determinados por SDS-PAGE, y de antígenos lipopolisacáridos estudiados por inmunoblot, de las cepas de referencia y de campo de los serovares 8 y 15 fueron similares entre sí. El análisis genético (16S rDNA, apxIIA, apxIIA, cps, genes cpx, apx y los perfiles omlA) reveló que los genes apxIIA y apxIIIA de las cepas de campo de los serovares 8 y 15 fueron similares a sus homólogos de las cepas de referencia de los serovares 3, 4, 6, 8 y 15. Los resultados obtenidos en el presente estudio pueden ser útiles para el desarrollo de vacunas más efectivas contra la enfermedad causada por A. pleuropneumoniae, al posibilitar incluir antígenos homólogos a los serovares prevalentes en las áreas geográficas de interés.
Actinobacillus pleuropneumoniae is the causative agent of porcine pleuropneumonia, an economically important bacterial disease of swine7. The virulence of A. pleuropneumoniae has been linked with exotoxins, capsular polysaccharides (CPS), lipopolysaccharides (LPS) and membrane proteins5,7. To date, 15 serovars have been recognized mainly on the basis of the antigenic properties of CPS and the O-polysaccharide (O-PS)3,5,24,25, and another one, serovar 16, was proposed based on serology alone26.
It has been shown that the characterization of the A. pleuropneumoniae serovar involved is useful for understanding the epidemiology of an outbreak, for preparing vaccines for the control of the disease and for serological monitoring of infected herds7. Cross-reactions between some serovars (1, 9 and 11; 4 and 7; and 3, 6, 8 and 15) and variable results between individual batches of test sera are usually observed in conventional serotyping tests7–9,13,14. To overcome these limitations, PCR assays based on the capsule loci have been developed for serotyping of A. pleuropneumoniae strains4,31,32.
Currently, numerous researchers have identified serovar 8 or 15 within the 3-6-8-15 cross-reacting group3,5,9,12,16,17,21,23,31. An understanding of the basic characteristics of strains of serovar 8 or 15 may be critical for the design of a vaccine against the infection by serovars belonging to this cross-reacting group. The aims of this work were to determine the serovars of A. pleuropneumoniae strains belonging to the 3-6-8-15 cross-reacting group isolated from pneumonic lesions of naturally infected dead or diseased pigs in Japan and Argentina. The serovar-identified strains were then characterized phenotypically and genetically by different techniques and compared to reference strains.
Materials and methodsBacterial strains and culture conditionsThe bacterial strains used in this study included nine reference strains of A. pleuropneumoniae (4074, serovar 1; CCM5870, 2; S1421, 3; M62, 4; K17, 5a; Femo, 6; 405 and CCM3803, serovar 8 and HS 143, 15), seventeen strains randomly chosen from 35 strains isolated from pneumonic naturally infected dead or diseased pigs in Argentina and 30 strains from pigs suffering from acute pleuropneumonia in Japan. The source of the field strains of A. pleuropneumoniae is shown in Table 1. The initial analyses revealed that these field strains possess the following toxin-related genes: apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA. The characteristics of some Argentine and Japanese strains were reported in a Master's thesis by Fernando Bessone at the University of Buenos Aires, Argentina in 20122 and at the 4th Asian Pig Veterinary Society Congress in Tsukuba, Japan, in 2009, respectively. In addition to the reference strains, four field strains, each from Japan and Argentina, were randomly chosen for the phenotypic and genetic analyses.
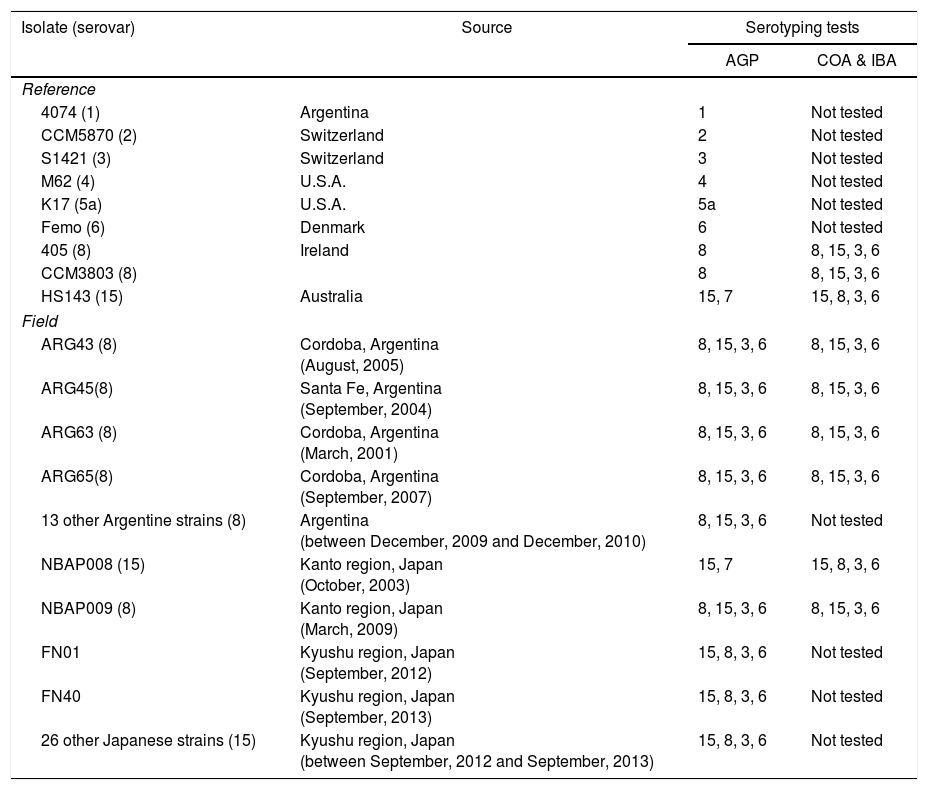
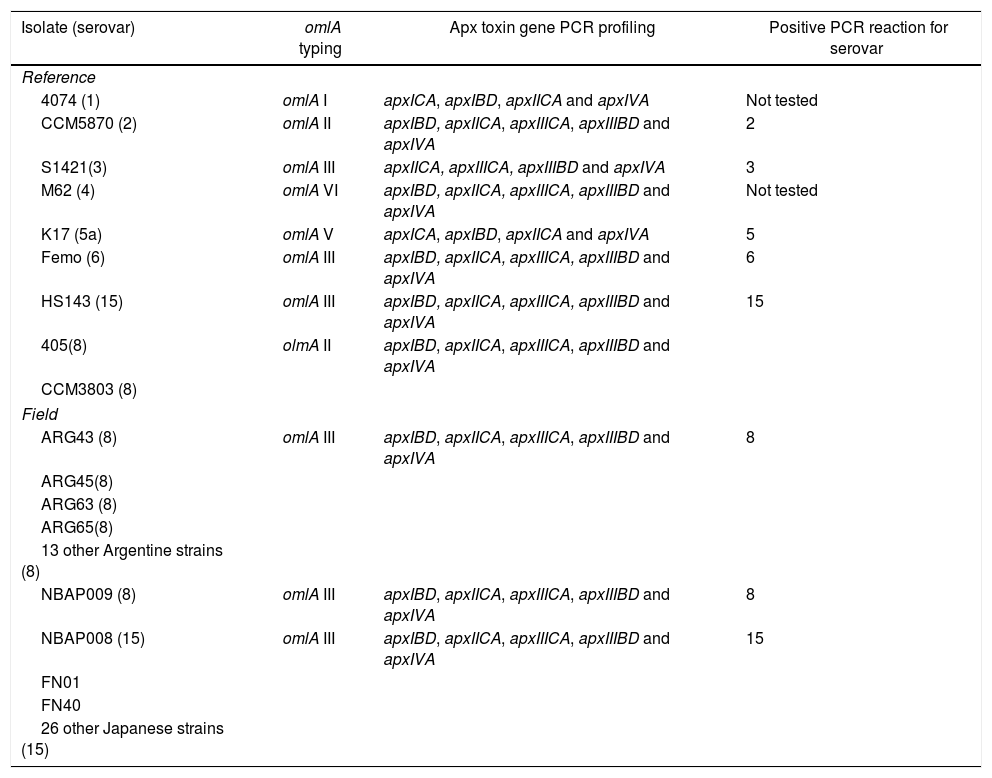
Source and some serological characteristics of Actinobacillus pleuropneumoniae strains of serovars 8 and 15
| Isolate (serovar) | Source | Serotyping tests | |
|---|---|---|---|
| AGP | COA & IBA | ||
| Reference | |||
| 4074 (1) | Argentina | 1 | Not tested |
| CCM5870 (2) | Switzerland | 2 | Not tested |
| S1421 (3) | Switzerland | 3 | Not tested |
| M62 (4) | U.S.A. | 4 | Not tested |
| K17 (5a) | U.S.A. | 5a | Not tested |
| Femo (6) | Denmark | 6 | Not tested |
| 405 (8) | Ireland | 8 | 8, 15, 3, 6 |
| CCM3803 (8) | 8 | 8, 15, 3, 6 | |
| HS143 (15) | Australia | 15, 7 | 15, 8, 3, 6 |
| Field | |||
| ARG43 (8) | Cordoba, Argentina (August, 2005) | 8, 15, 3, 6 | 8, 15, 3, 6 |
| ARG45(8) | Santa Fe, Argentina (September, 2004) | 8, 15, 3, 6 | 8, 15, 3, 6 |
| ARG63 (8) | Cordoba, Argentina (March, 2001) | 8, 15, 3, 6 | 8, 15, 3, 6 |
| ARG65(8) | Cordoba, Argentina (September, 2007) | 8, 15, 3, 6 | 8, 15, 3, 6 |
| 13 other Argentine strains (8) | Argentina (between December, 2009 and December, 2010) | 8, 15, 3, 6 | Not tested |
| NBAP008 (15) | Kanto region, Japan (October, 2003) | 15, 7 | 15, 8, 3, 6 |
| NBAP009 (8) | Kanto region, Japan (March, 2009) | 8, 15, 3, 6 | 8, 15, 3, 6 |
| FN01 | Kyushu region, Japan (September, 2012) | 15, 8, 3, 6 | Not tested |
| FN40 | Kyushu region, Japan (September, 2013) | 15, 8, 3, 6 | Not tested |
| 26 other Japanese strains (15) | Kyushu region, Japan (between September, 2012 and September, 2013) | 15, 8, 3, 6 | Not tested |
AGP: reactions by agar gel precipitation; COA: coagglutination assay; IBA: immunoblotting assay.
Actinobacillus pleuropneumoniae strains were cultured in chocolate agar (BD, Becton, Dickinson Co., Detroit, MI, USA) or in heart infusion medium (BBL, Cockeysville, MD, USA) supplemented with 0.3% yeast extract (dried yeast extract-S, Nippon Seiyaku, Tokyo, Japan) and 0.005% β-nicotinamide adenine dinucleotide (Oriental Yeast Tokyo, Japan). Escherichia coli was grown in LB medium. When appropriate, the LB medium was supplemented with ampicillin (50μg/ml).
Serological characterizationThe immunological-based serotyping was carried out by an agar gel precipitation test (AGP) and coagglutination test (COA) using rabbit antisera against serovars 3, 6, 8 and 15 of A. pleuropneumoniae20.
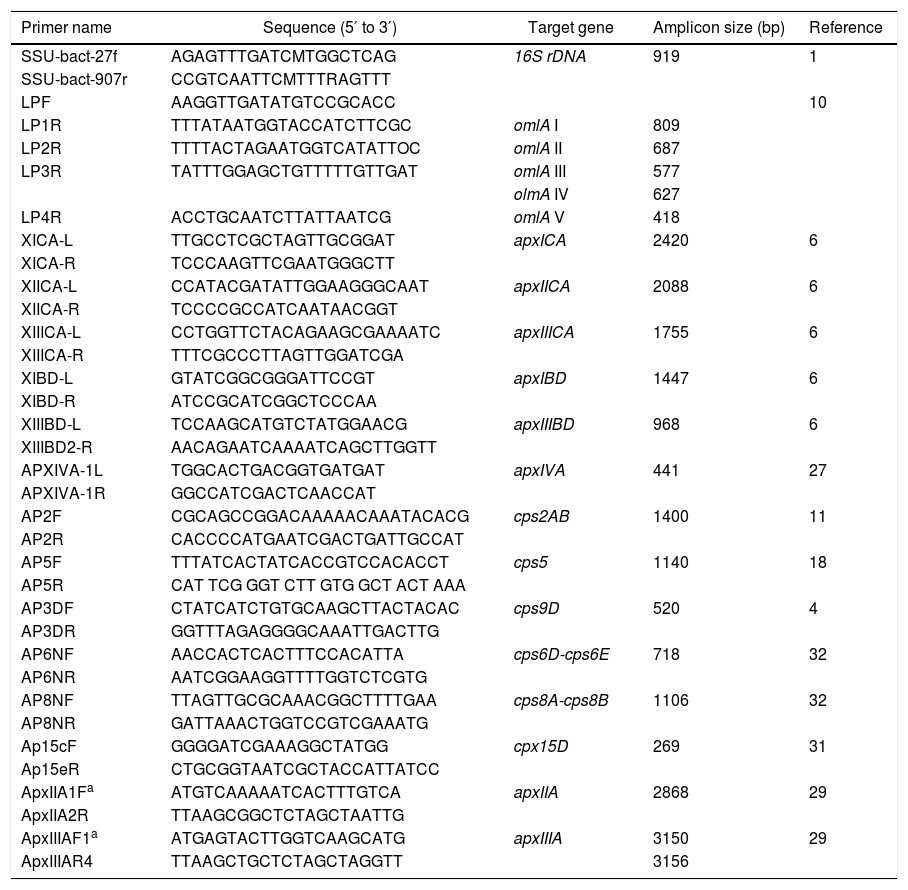
PCR detection of the apx and omlA genesApx toxin gene PCR profiling (detecting the presence of apxICA, apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA) and outer membrane lipoprotein (omlA) PCR typing were examined as previously described6,10,27. The primers used in this study are detailed in Table 3.
PCR, cloning and sequencing of cps, cpx, 16S rDNA and apxA genesThe serovars of field isolates were determined using PCR based on capsule loci4,11,18,31,32. Capsule loci (cps or cpx gene) typing was done as four single-reaction PCRs, one each for serovars 3, 6, 8 and 15, based on existing multiplex PCRs4,31,32. A 919 bp DNA fragment encoding a hypervariable region of 16 rRNA was PCR-amplified1. The PCR products from the 16S rRNA, cps and cpx genes were cleaned up using ExoSAP-I (Amersham Pharmacia Biotech, Uppsala, Sweden) and sequenced. To determine the nucleotide sequence of the open reading frame (ORF) encoding apxIIA or apxIIIA, the amplified DNA fragments encoding the full-length of apxIIA or apxIIIA were inserted into plasmid pGEM-T Easy (Promega, Madison, WI) and transformed into Escherichia coli XL1-Blue by electroporation29 DNA sequences from recombinant plasmids were determined by a primer-walking procedure29. In addition, phylogenetic relationships were conducted by using MEGA version 6.
Extraction of LPS and CPSFractions containing both LPS and CPS were extracted from bacterial cells of the five reference strains (HS143, serovar 15; 405, 8; 4074, 1; CCM5870, 2 and K17, 5a) and six field strains (NBAP008 and FN01, serovar 15; NBAP009, ARG65, ARG43 and ARG45, serovar 8) by the lysozyme–phenol–water method15.
SDS–PAGE and immunoblottingOvernight cultures of three reference strains (HS143, serovar 15; CCM 3803 and 405 serovar 8), and six field strains (NBAP008 and FN01, serovar 15; NBAP009, ARG65, ARG43 and ARG45, serovar 8) were used to prepare whole-cell lysates for protein analysis. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting were performed as described elsewhere28,29. The gels containing proteins were stained with Coomassie brilliant blue R250 (CBB), whereas the gels containing LPS were stained with silver as previously described28. Antigenic LPSs electroblotted onto a polyvinylidene difluoride (PVDF) membrane were probed with rabbit antisera against the various serovars, and with pig sera collected one week after challenge with strains HS143 or NBAP008 of serovar 15 and visualized with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins (Zymed Laboratories, Inc., San Francisco, CA, USA) or horseradish peroxidase-conjugated goat anti-swine IgG (Rockland Immunochemicals Inc., PA, U.S.A.) and a substrate solution containing 4-chloro-1-naphthol and hydrogen peroxide.
Nucleotide sequence accession numbersThe partial 16S rRNA sequences of the Argentine strains (ARG43, ARG45, ARG63, ARG65) and Japanese strains (NBAP009 and NBAP008) were submitted to the DDBJ/GenBank/EMBL databases under accession numbers. LC032461–LC032466. The apxIIA and apxIIIA sequences of the field strains (ARG43, ARG45, ARG63, ARG65, NBAP009 and NBAP008) and the reference strains (CCM3803, S1421, M62 and HS143) were deposited into the DDBJ/GenBank/EMBL databases under accession numbers LC043181–LC034187 and LC033888–LC033894, respectively. The nucleotide sequences of 292 bp fragment of serovar 15-specific cpx gene and of 1106 bp fragment of serovar 8-specific cps gene were submitted to the GenBank databases under accession numbers LC033493 and LC032467.
ResultsSerotyping by immunological-based methodsIn the AGP test, these strains tested displayed a clear line of precipitation to rabbit antiserum against serovar 8 or 15 and faint lines of precipitation to antisera against serovars 3, 6, 8, 15 or 7. These strains also showed a clear reaction by the COA with rabbit antisera against the reference strains of serovars 3, 6, 8 and 15 (Table 1).
Characterization of A. pleuropneumoniae strains by PCRThe PCR typing system based on the apx genes of six reference strains (CCM5870, serovar 2; S1421, 3; M62, 4; Femo, 6; HS143, 15; 405 and CCM3803, serovar 8) and forty-seven field strains showed the presence of apxIBD (genes for secretion of ApxI), apxIICA and apxIIICA (genes for expression and activation of ApxII and ApxIII, respectively), apxIIIBD (genes for secretion of ApxIII) and apxIVA (gene for expression of ApxIV) (Table 2).
Some genetic characteristics of Actinobacillus pleuropneumoniae strains of serovars 8 and 15
| Isolate (serovar) | omlA typing | Apx toxin gene PCR profiling | Positive PCR reaction for serovar |
|---|---|---|---|
| Reference | |||
| 4074 (1) | omlA I | apxICA, apxIBD, apxIICA and apxIVA | Not tested |
| CCM5870 (2) | omlA II | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | 2 |
| S1421(3) | omlA III | apxIICA, apxIIICA, apxIIIBD and apxIVA | 3 |
| M62 (4) | omlA VI | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | Not tested |
| K17 (5a) | omlA V | apxICA, apxIBD, apxIICA and apxIVA | 5 |
| Femo (6) | omlA III | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | 6 |
| HS143 (15) | omlA III | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | 15 |
| 405(8) | olmA II | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | |
| CCM3803 (8) | |||
| Field | |||
| ARG43 (8) | omlA III | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | 8 |
| ARG45(8) | |||
| ARG63 (8) | |||
| ARG65(8) | |||
| 13 other Argentine strains (8) | |||
| NBAP009 (8) | omlA III | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | 8 |
| NBAP008 (15) | omlA III | apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA | 15 |
| FN01 | |||
| FN40 | |||
| 26 other Japanese strains (15) | |||
The results of the PCR olmA gene typing showed that the field strains tested produced an omlA III PCR product (577bp) identical to that seen in the reference strains of serovars 3, 6 and 15, while reference strains of serovar 8 (405 and CCM3803) produced an omlA II PCR product (687bp). The reaction patterns of both the PCRs for the reference and field strains are detailed in Table 2.
Serotyping by CPS-specific gene-based PCRsThe fragment of approximately 292bp of cpx gene of serovar 15 was amplified from reference strain HS143 of serovar 15 and 29 Japanese strains. Whereas, the fragment of approximately 1106bp (cps8a-cps8B) of cps gene of serovar 8 was amplified from the reference strains of serovar 8 (405 and CCM3803), 17 Argentine strains and 1 Japanese strain. Serovar-specific primers for the reference strains of serovars 2, 3, 5 and 6 PCR amplified cps- or cpx-specific targets from the 4 reference strains (Table 2).
The nucleotide sequence similarity of the 292 bp fragment amplified from strains HS143 and from the 29 Japanese strains of serovar 15 was 100%. The sequence similarity of the 1106 bp (cps8A-cps8B) fragment amplified from reference strains 405 and CCM3803, 5 representative field strains (ARG43, ARG45, ARG63, ARG65, NBAP009) and other 13 field strains of serovar 8 was 100%.
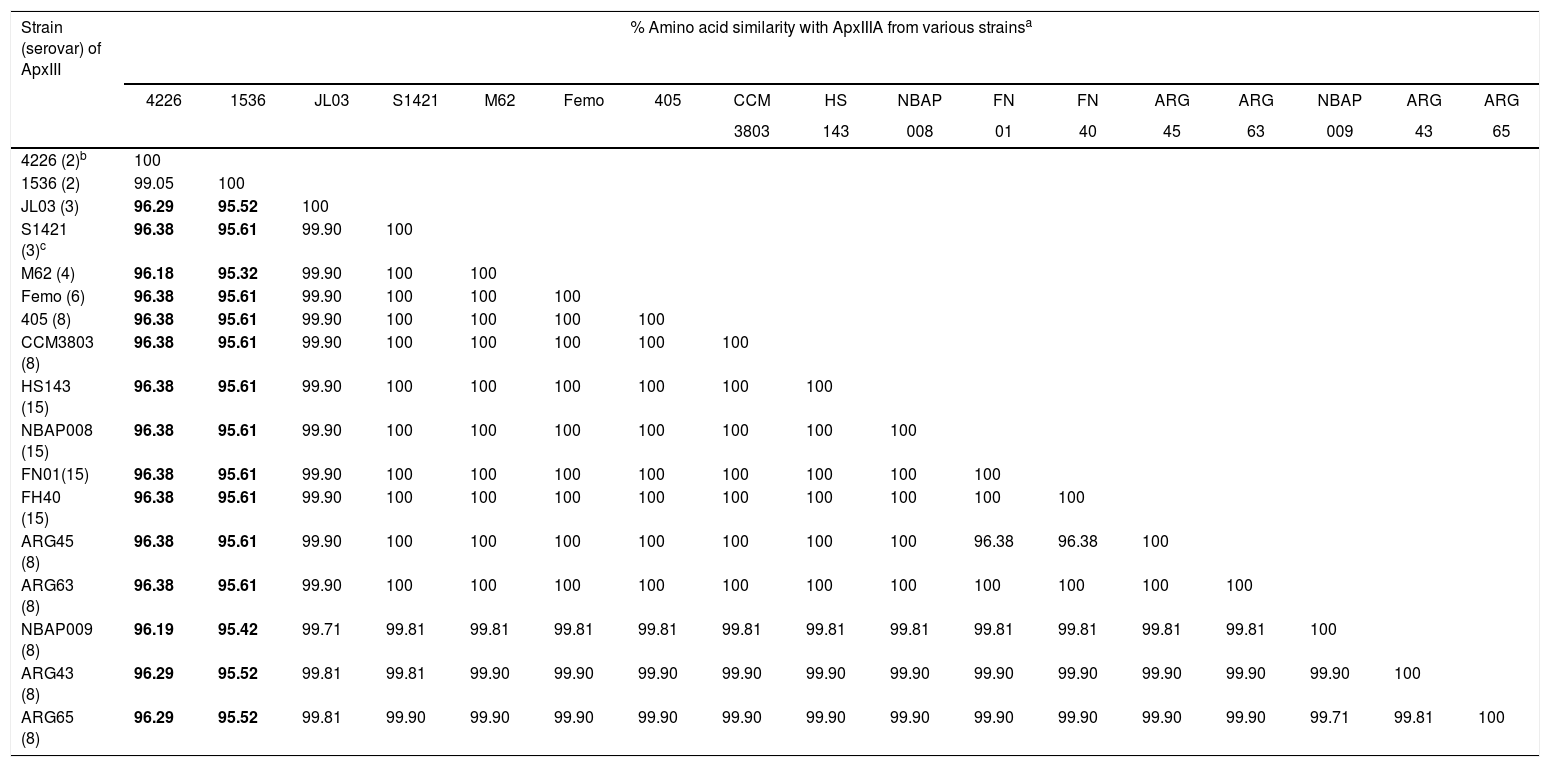
Sequence analysis of the apxIIA and apxIIIA genesThe sequence analysis revealed that the apxIIA ORF of eleven strains tested, including three reference strains (405 and CCM3803, serovar 8 and HS143, serovar 15), four Argentine strains of serovar 8 (ARG43, ARG45, ARG63 and ARG65) and four Japanese strains (NBAP008, FN01 and FN40, serovar 15 and NBAP009, serovar 8) was 2868bp in size (encoding 956 aa) similar to that of serovars 1, 2, 3, 4, 5, 6, 7, 9, 11, 12 and 13. The amino acid sequence similarities of ApxIIA among various serovars were 99.4–100% (ApxIIA sequences of serovars 1, 2, 3, 5, 6, 7, 9, 11, 12 and 13 were obtained from the GenBank under numbers EFM85442.1, EFL77686.1, ABY69525, ABN74052.1, EFL80296.1, ACE61660.1, EFM94174.1, EFM98503.1, EFN00640.1 and EFN02774.1, respectively). The size of apxIIIA ORF of the representative field strains tested of serovars 8 and 15 was 3156bp (encoding 1052 aa), similar to that of reference strains of serovars 3, 4, 6, 8 and 15, but different from those of strains of serovar 2 (3147bp for reference strain 1536 and 3150bp for Danish strain 4226, encoding 1049 and 1050 aa, respectively) (Table 4).
Primers used in this study
| Primer name | Sequence (5′ to 3′) | Target gene | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| SSU-bact-27f | AGAGTTTGATCMTGGCTCAG | 16S rDNA | 919 | 1 |
| SSU-bact-907r | CCGTCAATTCMTTTRAGTTT | |||
| LPF | AAGGTTGATATGTCCGCACC | 10 | ||
| LP1R | TTTATAATGGTACCATCTTCGC | omlA I | 809 | |
| LP2R | TTTTACTAGAATGGTCATATTOC | omlA II | 687 | |
| LP3R | TATTTGGAGCTGTTTTTGTTGAT | omlA III | 577 | |
| olmA IV | 627 | |||
| LP4R | ACCTGCAATCTTATTAATCG | omlA V | 418 | |
| XICA-L | TTGCCTCGCTAGTTGCGGAT | apxICA | 2420 | 6 |
| XICA-R | TCCCAAGTTCGAATGGGCTT | |||
| XIICA-L | CCATACGATATTGGAAGGGCAAT | apxIICA | 2088 | 6 |
| XIICA-R | TCCCCGCCATCAATAACGGT | |||
| XIIICA-L | CCTGGTTCTACAGAAGCGAAAATC | apxIIICA | 1755 | 6 |
| XIIICA-R | TTTCGCCCTTAGTTGGATCGA | |||
| XIBD-L | GTATCGGCGGGATTCCGT | apxIBD | 1447 | 6 |
| XIBD-R | ATCCGCATCGGCTCCCAA | |||
| XIIIBD-L | TCCAAGCATGTCTATGGAACG | apxIIIBD | 968 | 6 |
| XIIIBD2-R | AACAGAATCAAAATCAGCTTGGTT | |||
| APXIVA-1L | TGGCACTGACGGTGATGAT | apxIVA | 441 | 27 |
| APXIVA-1R | GGCCATCGACTCAACCAT | |||
| AP2F | CGCAGCCGGACAAAAACAAATACACG | cps2AB | 1400 | 11 |
| AP2R | CACCCCATGAATCGACTGATTGCCAT | |||
| AP5F | TTTATCACTATCACCGTCCACACCT | cps5 | 1140 | 18 |
| AP5R | CAT TCG GGT CTT GTG GCT ACT AAA | |||
| AP3DF | CTATCATCTGTGCAAGCTTACTACAC | cps9D | 520 | 4 |
| AP3DR | GGTTTAGAGGGGCAAATTGACTTG | |||
| AP6NF | AACCACTCACTTTCCACATTA | cps6D-cps6E | 718 | 32 |
| AP6NR | AATCGGAAGGTTTTGGTCTCGTG | |||
| AP8NF | TTAGTTGCGCAAACGGCTTTTGAA | cps8A-cps8B | 1106 | 32 |
| AP8NR | GATTAAACTGGTCCGTCGAAATG | |||
| Ap15cF | GGGGATCGAAAGGCTATGG | cpx15D | 269 | 31 |
| Ap15eR | CTGCGGTAATCGCTACCATTATCC | |||
| ApxIIA1Fa | ATGTCAAAAATCACTTTGTCA | apxIIA | 2868 | 29 |
| ApxIIA2R | TTAAGCGGCTCTAGCTAATTG | |||
| ApxIIIAF1a | ATGAGTACTTGGTCAAGCATG | apxIIIA | 3150 | 29 |
| ApxIIIAR4 | TTAAGCTGCTCTAGCTAGGTT | 3156 |
Deduced amino acid sequence similarities among ApxIIIA from 17 Actinobacillus pleuropneumoniae strains
| Strain (serovar) of ApxIII | % Amino acid similarity with ApxIIIA from various strainsa | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4226 | 1536 | JL03 | S1421 | M62 | Femo | 405 | CCM | HS | NBAP | FN | FN | ARG | ARG | NBAP | ARG | ARG | |
| 3803 | 143 | 008 | 01 | 40 | 45 | 63 | 009 | 43 | 65 | ||||||||
| 4226 (2)b | 100 | ||||||||||||||||
| 1536 (2) | 99.05 | 100 | |||||||||||||||
| JL03 (3) | 96.29 | 95.52 | 100 | ||||||||||||||
| S1421 (3)c | 96.38 | 95.61 | 99.90 | 100 | |||||||||||||
| M62 (4) | 96.18 | 95.32 | 99.90 | 100 | 100 | ||||||||||||
| Femo (6) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | |||||||||||
| 405 (8) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | ||||||||||
| CCM3803 (8) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | |||||||||
| HS143 (15) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | 100 | ||||||||
| NBAP008 (15) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||||||
| FN01(15) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | ||||||
| FH40 (15) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||||
| ARG45 (8) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 96.38 | 96.38 | 100 | ||||
| ARG63 (8) | 96.38 | 95.61 | 99.90 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||
| NBAP009 (8) | 96.19 | 95.42 | 99.71 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 99.81 | 100 | ||
| ARG43 (8) | 96.29 | 95.52 | 99.81 | 99.81 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 100 | |
| ARG65 (8) | 96.29 | 95.52 | 99.81 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.90 | 99.71 | 99.81 | 100 |
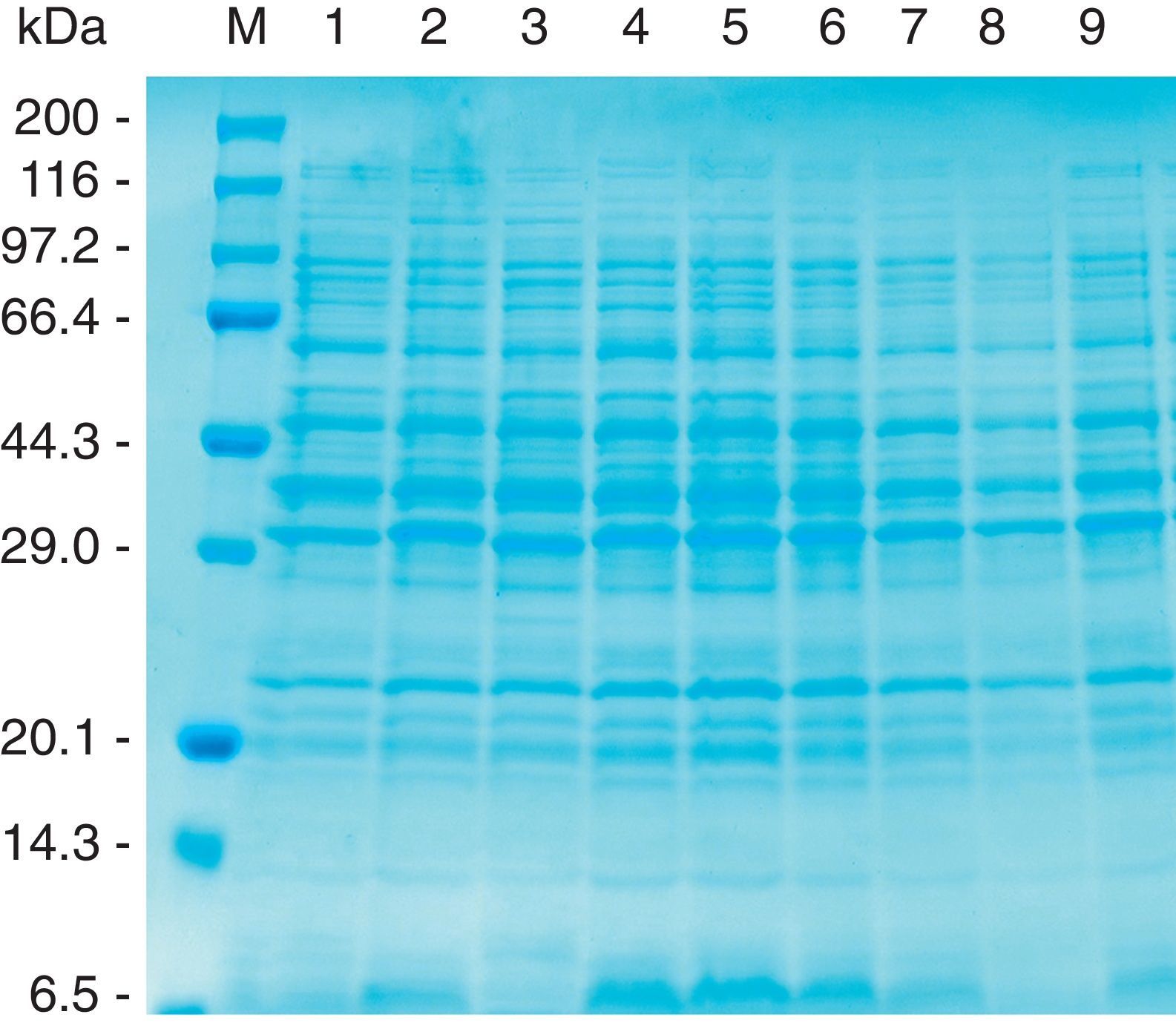
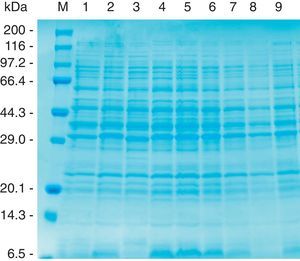
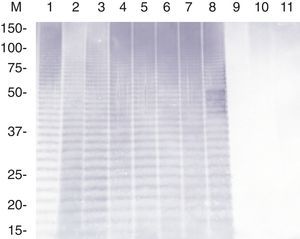
The CBB-stained SDS–PAGE patterns of whole-cell proteins of three reference strains (HS143, serovar 15; CCM 3803 and 405 serovar 8), and six field strains (NBAP008 and FN01, serovar 15; NBAP009, ARG65, ARG43 and ARG45, serovar 8) are shown in Figure 1. Six major polypeptides (approximately 96, 60, 46, 40, 32 and 23kDa) were found to be common to the reference and field strains of serovars 8 and 15. The polypeptide patterns of A. pleuropneumoniae strains were similar to each other, with minor differences in the range below 14.3kDa.
Whole-cell protein patterns of A. pleuropneumoniae strains of serovars 8 and 15 on 12.5% polyacrylamide gel stained with CBB. Lanes: 1, HS143 (serovar 15); 2, NBAP008 (serovar 15); 3, CCM 3803 (serovar 8); 4, FN01 (serovar 15); 5, NBAP009 (serovar 8): 6, ARG65 (serovar 8); 7, ARG43 (serovar 8); 8, ARG45 (serovar 8); and 9, 405 (serovar 8). M, molecular weight marker in kilodaltons (kDa).
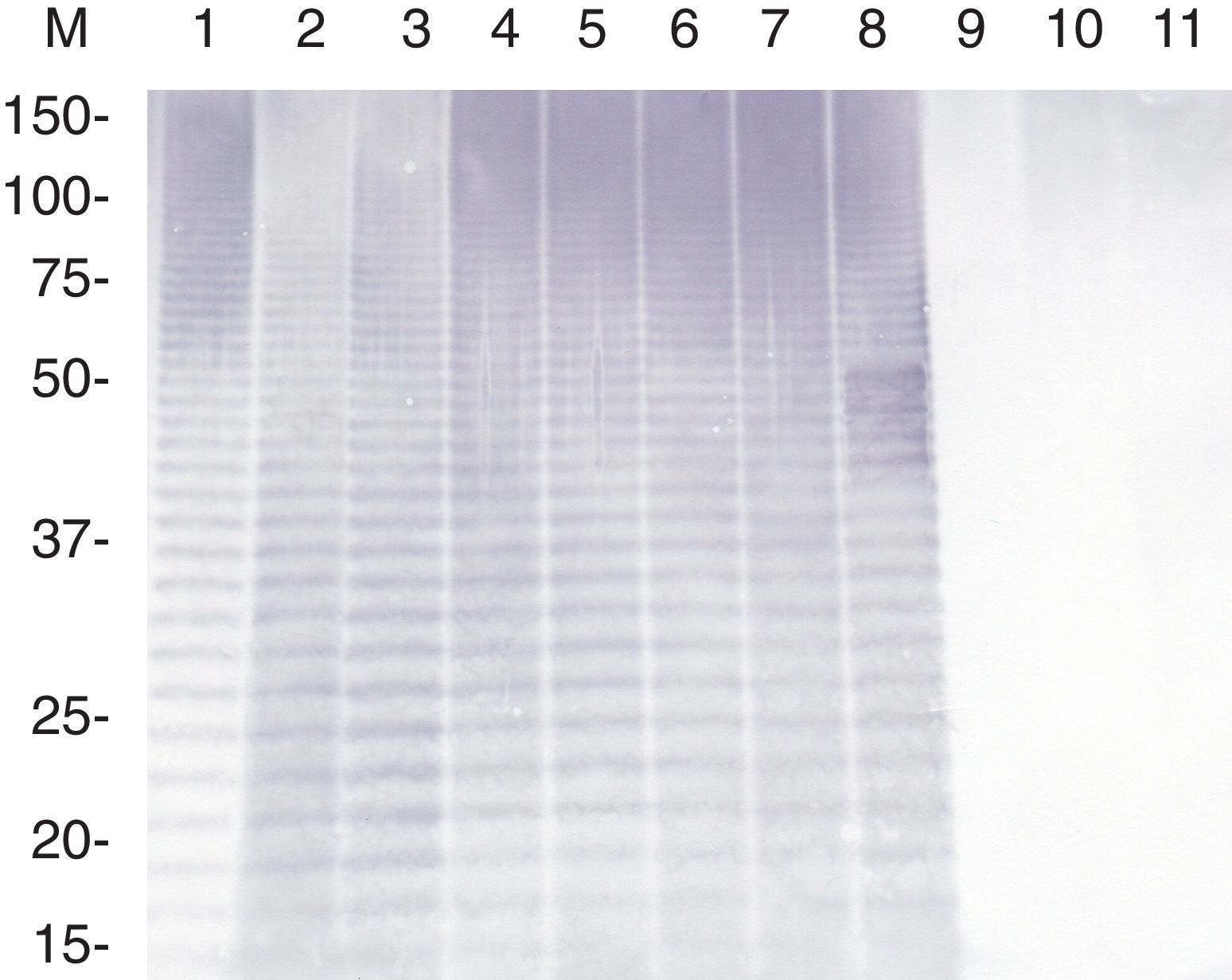
A high-molecular-mass area of long-chain lipopolysaccharide and a ladder-like pattern typical of smooth LPS of A. pleuropneumoniae were observed in gels stained with silver, and in the immunoblots probed with rabbit antisera raised against A. pleuropneumoniae strains HS143 and NBAP008 or with pig sera collected one week after challenge with strains HS143 and NBAP008. Immunoblotting analysis revealed that LPS of the reference and field strains (HS143 and NBAP008) of serovar 15, and of the reference (405) and field strains (ARG43, ARG45, ARG63, ARG65, NBAP009) of serovar 8 reacted strongly not only with rabbit antisera against serovars 3, 6, 8, 15 (Table 1) but also with sera derived from serovar 15-infected pigs (Fig. 2).
Immunoblot analysis of the purified LPS of A. pleuropneumoniae strains of serovars 8 and 15 probed with sera derived from pigs experimentally infected with reference strain HS143 of serovar 15. Lanes: 1, HS143 (serovar 15); 2, 405 (serovar 8); 3, NBAP008 (serovar 15); 4, FN01 (serovar 15); 5, NBAP009 (serovar 8): 6, ARG65 (serovar 8); 7, ARG43 (serovar 8); 8, ARG45 (serovar 8); 9, 4074 (serovar 1); 10, CCM5870 (serovar 2); and 11, K17 (serovar 5a). M, molecular weight marker in kilodaltons.
Similarity analysis of the partial 16S rRNA sequences of the A. pleuropneumoniae strains showed homology ranging from 99.2 to 99.8% with those of other A. pleuropneumonia serovars available in the GenBank database.
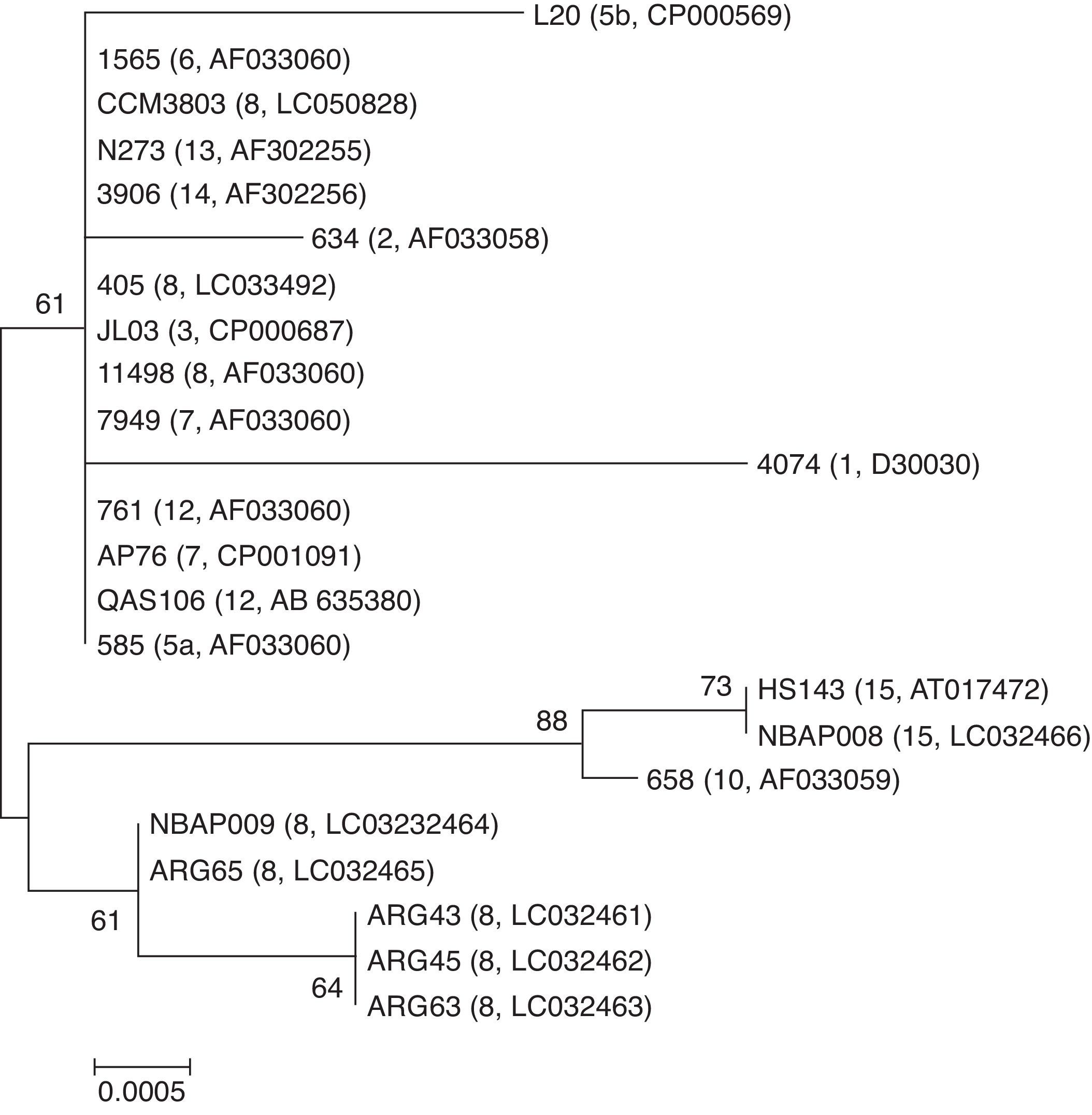
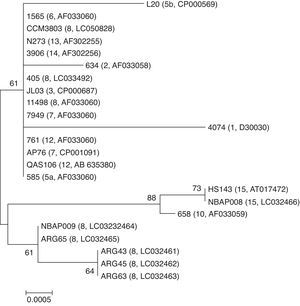
In the phylogenetic analysis of a 919 bp hypervariable region of 16S rRNA gene, a group of the field strains of serovar 8 clustered closest to a group of strains of serovar 15 (Fig. 3). In contrast, serovar 8 reference strains (405 and CCM3803) and Danish field strain 11498 clustered in a different group including serovars 3, 5, 6, 7, 12, 13 and 14.
Neighbor-joining phylogenetic tree based on the partial nucleotide sequences of 16S rRNA gene of 23 A. pleuropneumoniae strains (serovars and accession numbers are given in brackets). The tree was constructed by using MEGA version 6. Bootstrap values of 500 simulations are shown at the branches. The field strains mentioned in the present study are shown in bold. The bar with a number indicates genetic distance.
The forty-seven field strains (including 30 Japanese strains and 17 Argentine strains) were considered to belong to the 3-6-8-15 cross-reacting group by the immunological-based method. The capsule loci-based typing revealed that eighteen and twenty-nine strains were serovars 8 and 15, respectively. By traditional serological methods, the final identification of the serovars within the 3-6-8-15 cross-reacting group is extremely difficult5,7–9,13,23. It has been known that cross-reactions between serovars 3, 6, 8 and 15 could be due to common epitopes located on the LPS O-chains5,8,9,24,25. Our observation that conventional serotyping tests in combination with serovar-specific PCR could identify the actual serovar of isolates within 3-6-8-15 cross-reacting group is consistent with data from previous reports9,17,23. The finding concerning the serological characterization results underlines the significance of capsule loci-based PCR when the immunological-based serotyping seems questionable.
It is also worth noting that the sequence analysis of the cpx fragment from strain HS143 and 29 field strains of serovar 15 revealed that the size of the PCR product was 292bp instead of 269bp as reported by Turni et al.31 The nucleotide sequence analyses of the 292 bp fragments amplified from strains HS143 and from the 29 Japanese A. pleuropneumoniae strains of serovar 15 support the results mentioned on the size of the PCR product of serovar 15 presented in this study.
The serovar prevalence among strains recovered from diseased animals was reported to vary depending on the continent and time5,8. In a review of seroprevalence around the world, 25 of 27 countries reported the presence of serovars 3, 6 or 85. Updating epidemiological data is thought to be important since the distribution of serovars within a country may change over time. For example, serovars 5 and 7 currently dominate in Canada9, whereas until the 1990s, it was serovar 18. Recently, several investigations showed that serovars 8 and 15 are highly prevalent among diseased pigs in UK17,23 and Australia3,31, respectively. In addition, serovar 8 was also found to be the predominant serovar within the 3-6-8-15 cross-reacting group in North America8,9. Similar to what has been reported in North America, the current study found that serovars 8 and 15 were prevalent serovars within the 3-6-8-15 cross-reacting group in various parts of Argentina and Japan, respectively. In Argentina, a prevalence study of A. pleuropneumoniae clinical isolates collected from diseased pigs in 19885 by an immunological-based method (COA) showed that serovar 3 was the predominant serovar within the 3-6-8-15 cross-reacting group. The presence of A. pleuropneumoniae isolates of serovar 8 in Argentina was found in 199633, and more recently, an investigation of A. pleuropneumoniae clinical isolates2 found 39% of isolates from a total collection of 131 Argentine strains belonging to the 3-6-8-15 group. Our results for the prevalence of serovars within the 3-6-8-15 cross-reacting group in Argentina were different from those reported previously5. The reasons for the discrepancy between their results and ours are not clear but may reflect variations in the course of time studied and serotyping method used, the immunological-based method (COA) versus the serovar-specific PCR method (based on CPS-specific genes). In addition, serovar switching over time has also been reported in several countries8. This is what could have happened in Argentina because of changes in swine genetics and production system. Our observation that serovar 15 is the predominant serovar within the 3-6-8-15 cross-reacting group, isolated from diseased pigs in Japan is consistent with data from previous reports by Morioka et al.21 and Ito12.
The toxin gene profile for the thirty Japanese and seventeen Argentine strains was uniform and showed the presence of apxIBD, apxIICA, apxIIICA, apxIIIBD and apxIVA, suggesting that the toxin gene pattern of these strains was typical of A. pleuropneumoniae serovars 2, 4, 6, 8 and 153, because strains of serovar 3 lack apxIBD genes6.
The results showed that the omlA genes of the field strains of serovar 8 from Argentina and Japan are different from those of the reference strains. Data from a previous report10 showed that the omlA genes of A. pleuropneumoniae strains corresponded to their serovars, with the exception of divergence in the omlA gene between the reference strains and the Danish field strains of serovar 8. The present finding concerning divergence of the omlA gene of serovar 8 supports the previously reported results by Gram et al.10
The results of the PCR apx gene typing and the PCR omlA gene typing suggested that the field strains of A. pleuropneumoniae from Japan and Argentina belonged to serovars 6, 8 and 15.
The phylogenetic analysis based on a 919 bp hypervariable region of the 16S rRNA gene found that the field strains of serovar 8 are genetically similar to each other but different from the reference strains of serovar 8. The reasons for this difference are not known but may be due to differences in the geographical origin of the strains or differences between the recently isolated strains and the older ones, which were isolated more than twenty years ago.
The sequence analyses revealed that the apxIIA genes of the field and reference strains of serovars 8 and 15 were similar to those of the reference strains of the other serovars. Whereas the apxIIIA genes of the field strains of serovars 8 and 15 were similar to those of the reference strains of serovars 3, 4, 6, 8 and 15, suggesting that the apxIIIA gene is more preserved than the omlA gene of strains belonging to the 3-6-8-15 cross-reacting group.
The whole-cell protein patterns of the three reference strains (405, CCM 3803 and HS143), three Argentine strains (ARG43, AGR 45 and ARG 65) and three Japanese strains (NBAP009, NBAP008 and FN01) of serovars 8 and 15 were similar to each other by one-dimension SDS–PAGE, which is consistent with data from previous reports19,22. Those investigators found that the whole-cell protein patterns of A. pleuropneumoniae serovars 1–8 are very similar to each other. In SDS–PAGE and the immunoblotting analysis, LPS of the reference and field strains of serovars 8 and 15 were found to be antigenically similar to each other. The serological typing by AGP, COA and IBA revealed cross-reactions between LPS of the A. pleuropneumoniae strains tested with rabbit antisera against serovars 3, 6, 8 and 15. Additionally, the results of the serological typing together with those of the immunoblotting analysis (Fig. 2) suggested that the LPS of serovars 8 and 15 was antigenically different from those of serovars 1, 2 and 5, which was supported by data from previous reports5,7,9,24,25. The sequence analysis revealed that the apx genes of the field strains of serovars 8 and 15 were almost identical to those of the reference strains of serovars 3, 4, 6, 8 and 15, but different from those of serovar 2 at the carboxyl terminus, supporting results previously reported29. That investigation29 revealed that (i) the ApxIIA proteins of serovars 2, 3, 4, 6, 8 and 15 are genetically and antigenically very similar to those of the other A. pleuropneumoniae serovars and (ii) the apxIIIA proteins have two variants, one in strains of serovar 2 and the other in strains of serovars 3, 4, 6, 8 and 15.
In conclusion, the results of the present study revealed that (i) serovars 8 and 15 are prevalent ones within the 3-6-8-15 cross-reacting group in Argentina and Japan, respectively; and (ii) the lipopolysaccharide antigen and Apx toxins of the field strains of serovars 8 and 15 are similar to those of the reference strains of serovars 8 and 15. Commercially toxin-type vaccines have been shown to give high protection against a wide variety of A. pleuropneumoniae serovars7,30. However, a recent trial with one of these vaccines showed that the vaccine was unable to provide significant protection against strain HS143 (serovar 15) challenge30. Therefore, the present findings concerning antigenic similarities of LPS and Apx toxins of serovars 8 and 15 should be taken into consideration when designing vaccines for countries where isolates belonging to serovars 8 and/or 15 have been frequently found.
Authors’ contributionsFAB, HT, JL, KT and TK conducted the processing and analysis of samples in the laboratory, analyzed the data, and helped to draft the manuscript. AO, NT, SN and TN contributed to manuscript writing. HT and GCZ contributed to the design of the study and manuscript writing. All authors read and approved the final manuscript.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this study.
Confidentiality of dataThe authors declare that no patient data appear in this article.
Right to privacy and informed consentThe authors declare that no patient data appear in this article.
Conflicts of interestThe authors declare that they have no conflicts of interest.
The authors would like to thank Professor Pat Blackall (University of Queensland, Australia) for providing reference strain HS143, Professor Marcelo Gottschalk (University of Montreal, Canada) for providing A. pleuropneumoniae strains; Dr. Conny Turni (University of Queensland, Australia) and Emeritus Professor Chihiro Sasakawa (University of Tokyo) for their advice during preparation of the manuscript.