Allergen-specific immunotherapy (ASIT) is the only allergic disease-modifying therapy available for children and adults, and recombinant allergens are an interesting approach to improve allergy diagnosis and ASIT. Tyrophagus putrescentiae is a common storage mite that produces potent allergens. The aim of this study was to express and characterize recombinant group 4 allergen protein of T. putrescentiae (Tyr p 4), and to further investigate allergenicity and potential epitopes of Tyr p 4.
Materials and methodsThe cDNA encoding Tyr p 4 was generated by RT-PCR and subcloned into pET-28a(+) plasmid. The plasmid was then transformed into E. coli cells for expression. After purification by nickel affinity chromatography and identification by SDS-PAGE, recombinant Tyr p 4 protein was used for a skin prick test and an ELISA to determine the allergic response.
ResultsStudy participants’ allergic response rate to Tyr p 4 protein was 13.3% (16/120). Eight B-cell epitopes and three T-cell epitopes of Tyr p 4 were predicted.
ConclusionsSimilar to group 4 allergens of other species of mite, allergenicity of Tyr p 4 is weak. The expression, characterization and epitope prediction of recombinant Tyr p 4 protein provide a foundation for further study of this allergen in the diagnosis and ASIT of storage mite allergy.
Mites allergens are a common trigger of allergies in children and adults worldwide, and produce asthma, atopic dermatitis, and perennial rhinitis, among other symptoms.1 Allergens from house dust mites (HDMs), in particular those from the most common HDMs, Dermatophagoides farinae (Der f) and Dermatophagoides pteronyssinus (Der p), are major environmental factors for allergic diseases.2,3 However, many allergens from other mite species, such as Tyrophagus putrescentiae in storage mites, also contribute to allergic disease.4,5 Since there are currently few studies on allergens of T. putrescentiae, only eight groups of mite allergens from T. putrescentiae have been designated in the WHO/IUIS Allergen Nomenclature Database (http://www.allergen.org/). In addition to these eight groups of allergens, there are other allergens in T. putrescentiae that are not listed in the nomenclature database, like Tyr p 4, because their characterization and allergenicity are still unclear.
At present, treatment options for mite allergic diseases include allergen avoidance, pharmacotherapy, and allergen-specific immunotherapy (ASIT).6 Allergen avoidance and pharmacotherapy are currently the mainstay treatments of these diseases, but they are not easily able to change the course of disease.6 ASIT is the only allergic disease-modifying therapy available.7 However, ASIT with crude mite extracts, a mixture of allergens, nonallergens, and other proteins cannot be fully standardized and can cause severe side effects such as anaphylaxis.
Recombinant allergens are an interesting approach to improve allergy diagnosis and ASIT, because they can be modified to reduce allergenicity and retain immunogenicity, which may increase safety and efficacy of allergy treatments.8 Thus, researchers are working toward the development of recombinant allergens that mimic the native proteins and can be used for ASIT.8
Our laboratory has previously cloned and expressed several recombinant allergens from mites,9,10 which are expected to be used in ASIT in the future. Here, in order to better understand the allergens in T. putrescentiae and provide more recombinant allergens for ASIT, we sought to clone and express recombinant Tyr p 4 allergen protein, and then describe characterization, allergenicity and possible antigenic epitopes of Tyr p 4. These results provide a foundation for further study of this allergen in the diagnosis and ASIT of storage mite allergy.
Materials and methodsTyrophagus putrescentiae extractsTyrophagus putrescentiae extracts were supplied by Greer Laboratories (Lenoir, IL, USA) as a lyophilized powder from cleaned mites. It was dissolved in phosphate-buffered saline (PBS, pH 7.4), dialyzed against PBS, and diluted to a final concentration of 1g/L for skin prick test of serum provider.
Serum samplesSerum samples were obtained from 120 workers at five wheat milling factories in Xinghua City, Jiangsu Province, China. The 120 workers (aged 20–60 years, mean 36.57 years; 95 males and 25 females; mean working age, 3.98 years) were in close contact with wheat flour during work and all had allergic symptoms. Based on personal information and doctor's diagnosis, 89 of them had allergic rhinitis, 35 had mild asthma, and 28 had atopic dermatitis. All of them had positive skin reactions by the skin prick test (the ratio of the wheal area ≥++, the detailed steps are below) to T. putrescentiae extracts. None of these participants had undergone allergic treatment. Sera from 10 healthy adult volunteers with no history of any allergic symptom and a negative skin prick test response to common mite allergens were obtained as negative controls. All participants provided written informed consent. The study was approved by the Clinical and New Technology Ethics Committee of Wuxi People's Hospital (approval number is KYLLH2018034), Wuxi City, Jiangsu Province, China. Confidentiality is maintained.
Sequencing Tyr p 4Tyrophagus putrescentiae was cultured and total RNA was isolated as previously described.5 DNA primers were designed and synthesized based on the sequence of Tyr p 4 (GenBank accession no. DQ983318.1). The sequence of the forward primer was 5′-CATGCCATGGCTTTCAACAGCTACATTCTTC-3′ and the reverse primer was 5′-CCGCTCGATGCTTGGCATGCACGTGAATTG-3′. The reaction was performed as previously reported9: 2min at 98°C, 30 cycles of 30s at 98°C, 30s at 60°C, 10min at 72°C, and 5min at 10°C. 5μL of PCR product was analyzed by agarose gel electrophoresis (2.0%) and visualized with ImageMaster® VDS.
TA cloningA deoxyadenosine nucleotide (A) was added to the 3′ end of the PCR product using a Takara DNA A-tailing kit (Code No. 6109). The reaction mix of 50μL consisted of t10× A-Tailing Buffer (5μL), 2.5mM of dNTP Mixture (4μL), 5U/μL of A-tailing enzyme (0.5μL), smooth-ended DNA fragments (5μg), and ddH2O (35.5μL) and was incubated at 72°C for 20min before being placed on ice for 2min.
The A-tailed fragment was subjected to an enzymatic reaction with the pMD20T plasmid using NEB T4 DNA ligase (Code No. M0202). The reaction mixture of 10μL consisted of 10× T4 DNA ligase buffer (1μL), pMD20T vector DNA (4kb, 1μL), A-tail DNA fragments (6μL), T4 DNA ligase (0.5μL), and nuclease-free water (1.5μL) and was incubated at 4°C for 8h. E. coli JM109 (Takara Bio Inc., Code No. D9052) was then transformed with the recombinant plasmid pMD20T-Tyr p 4 and positive clones were selected by blue/white screening on Luria Bertani (LB) plates containing 100mg/mL ampicillin. The plasmid was then extracted for verification.
Construction of expression plasmid pET-28a(+)-Tyr p 4pMD20T-Tyr p 4 and pET-28a(+) were double digested. The 105-μL reaction mixture consisted of pMD20T-Tyr p 4 (50μL), 10× K buffer (5μL), 0.1% BSA (5μL), pET28a(+) plasmid (35μL), NcoI (2.5μL), XhoI (2.5μL), and DDH2O (5μL) and was incubated at 37°C for 6h. The product was then recovered by cutting fragments from the agarose gel. Then, 10× T4 DNA ligase buffer (1μL), pET-28a(+) plasmid recovered by double digestion (4kb, 2μL), Tyr p 4 recovered by double digestion (2μL), T4 DNA ligase (0.5μL), and nuclease-free water (4.5μL) were incubated at 4°C for 8h. E. coli DH5α competent cells (Takara Bio Inc., Code No. 9057) were transformed with the recombinant plasmid pET-28a(+)-Tyr p 4 and positive clones were selected by blue/white screening on Luria Bertani (LB) plates containing 50mg/mL kanamycin. The plasmid was then extracted for sequencing.
Production and purification of recombinant Tyr p 4 proteinThe pET-28a(+)-Tyr p 4 plasmids were transformed and expressed in E. coli BL21 (DE3, Stratagene, La Jolla, CA, USA), purified by nickel affinity chromatography, verified by SDS-PAGE and Western blotting, and analyzed by MALDI-TOF/TOF mass spectrometer (model 4800, Applied Biosystems) as previously described.10
Skin prick testParticipants were free of skin scratches and had no skin damage or infection at the test site. All stopped taking anti-allergy drugs and hormones for at least 3 days before the experiment. The skin of the inner forearm was cleaned with saline and the negative control solution (0.9% NaCl solution), the allergen solution (1g/L T. putrescentiae extracts or 1g/L recombinant Tyr p 4 protein), and the positive control solution (10g/L histamine phosphate solution) were sequentially applied to the skin of each participant. The distance between application sites was 3cm. Avoiding blood vessels, a needle was passed through the droplets to slightly puncture the skin without bleeding. The needle was removed after 1s, and the skin was observed after 15min. The ratio of the wheal area caused by T. putrescentiae extracts or recombinant Tyr p 4 protein liquid (S1) to the wheal area caused by positive control solution (S2) was determined. If S1/S2 was between 0 and 25% it was scored as negative (−), between 26% and 50% it was slightly positive (+), between 51% and 100% was positive (++), between 101 and 200% was (+++), and more than 200% was (++++). The result was judged to be positive at (++) or above.11
IgE-binding reactivity of recombinant Tyr p 4 proteinThe IgE-binding reactivity of Tyr p 4 allergen was detected by ELISA as previously described.9 Recombinant Tyr p 4 protein was coated onto a microplate, blocked in bovine serum albumin (BSA), and patient serum was added to the microtiter plate. Secondary antibody and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate solution were added to the microtiter plate to carry out the reaction. Finally, H2SO4 was added to terminate the reaction. The optical density was measured using an iMark microplate absorbance reader (Bio-Rad). Reactivity was considered positive if the optical density (OD) of the detected sera was higher than the cut-off ELISA value (within three standard deviations of the mean ELISA value of healthy donors). The specific IgE classes are defined using six calibrators: 0, 0.35, 0.7, 3.5, 17.5 and 100kUA/L (Class 0: from 0 to <0.35kUA/L; Class 1: from 0.35 to <0.7kUA/L; Class 2: from 0.70 to <3.5kUA/L; Class 3: from 3.50 to <17.5kUA/L; Class 4: from 17.5 to <50kUA/L; Class 5: from 50 to <100kUA/L; and Class 6: from ≥100kUA/L).12
Characterization of Tyr p 4 proteinThe amino acid sequence of Tyr p 4 protein (GenBank accession no. ABM53754.1) was obtained from the protein database of the National Center for Biotechnology Information (www.allergen.org). The open reading frame (ORF) was identified using ORF server.13 Transmembrane protein helices were predicted by TMHMM server 2.0.14 The physicochemical properties and hydrophilicity of the Tyr p 4 amino acid sequence was predicted using ProtParam,15 and phosphorylation sites were predicted using NetPhos 3.1 server.16 Phylogenetic tree construction and identification of Tyr p 4 amino acid sequence alignment were performed by MEGA 5.0217 and VECTOR NTI 11.0,18 respectively. Secondary structure was predicted using the JPred4 server.19
Structure prediction and homology modelingTemplates of Tyr p 4 protein were analyzed for homology modeling in the Protein Data Bank (PDB)20 using Protein BLAST. Based on high score, low e-value, and maximum sequence identity, the appropriate templates were selected. The tertiary structure of Tyr p 4 protein was predicted by homology modeling using MODELLER 9.22.21 PROCHECK determined the stereochemical quality of Tyr p 4 protein structure by analyzing residue-by-residue geometry and overall structural geometry.22 VERIFY 3D was used to detect the compatibility of an atomic model (3D) of Tyr p 4 protein with its own amino acid sequence (1D).23 ERRAT analyzed the statistics of non-bonded interactions between different atom types.24 ProSA was used to analyze the Z-score to determine the degree of match between the template and Tyr p 4 protein.25 QMEAN derived both global (the entire structure) and local (per residue) absolute quality estimates on the basis of one single model.26 Interactive visualization and analysis of molecular structures were completed using UCSF Chimera 1.13.1.27
Prediction of epitopesBcePred,28 ABCpred,29 and BCPreds30 were used to predict the B-cell antigenic epitopes of Tyr p 4. NetMHCpan 4.0,31 NetMHCII 2.3,32 and NetMHCIIpan 3.233 were used to predict the T-cell antigenic epitopes of Tyr p 4.
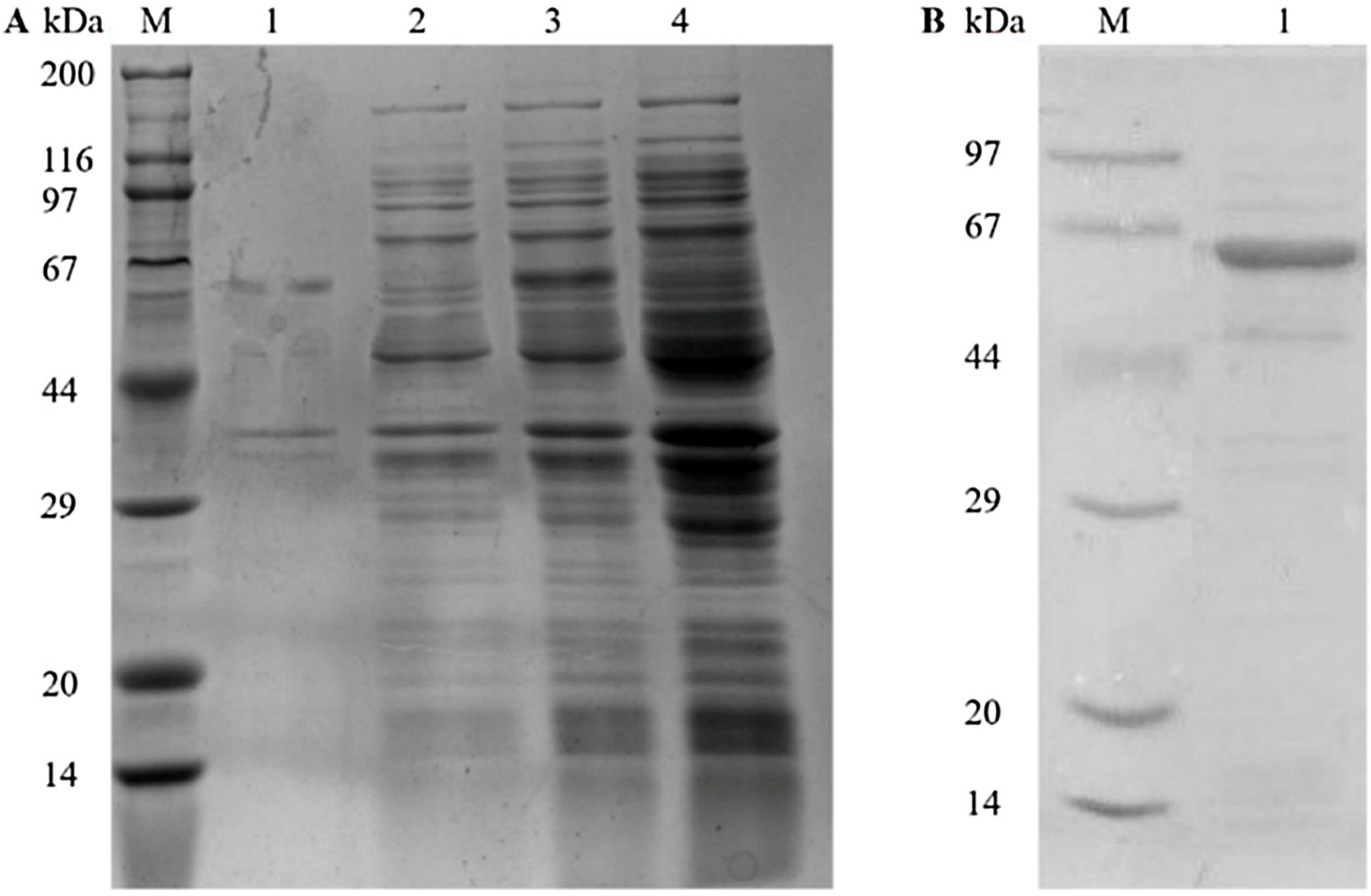
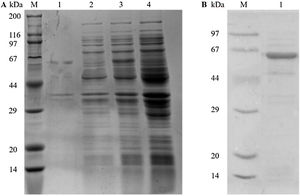
ResultsGene cloning, expression, and purification of recombinant Tyr p 4 proteinPlasmids from sequenced kanamycin-resistant clones were used to successfully transform pET-28a(+)-Tyr p 4 into E. coli DH5α cells and expressed under IPTG (Fig. 1A). A single and specific band matching the predicted molecular weight of Tyr p 4 using ProtParam was observed (Fig. 1B).
Expression and purification of rTyr p 4 protein. (A) SDS-PAGE demonstrating expression of rTyr p 4 protein in BL21 cells. Lane M, protein marker (TransGen Biotech); lane 1, the pellet of cells containing pET-28a(+)-Tyr p 4 3h after induction; lane 2, supernatant from cells containing pET-28a(+)-Tyr p 4 3h after induction; lane 3, whole BL21 cells containing pET-28b (+)-Tyr p 4 before IPTG induction; lane 4, whole BL21 cells containing pET-28a(+) before IPTG induction. (B) Tyr p 4 protein purified from BL21 cells. Lane M, protein marker (TransGen Biotech); lane 1, purified Tyr p 4 protein.
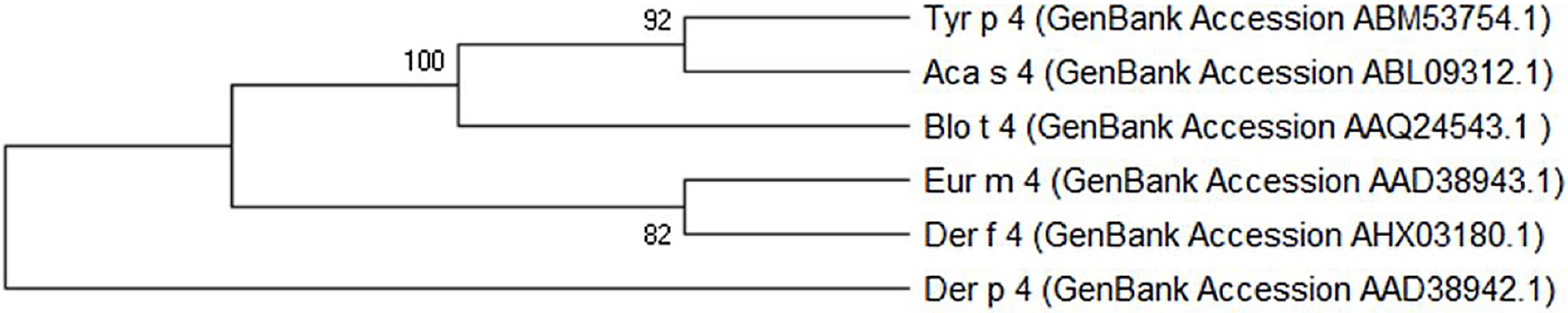
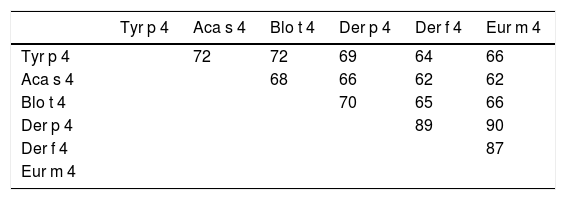
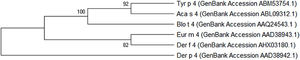
The Tyr p 4 gene contains a complete open reading frame, with 1557bp between the initiation codon ATG and the stop codon TAA, encoding a protein of 518 amino acids. Family classification shows that Tyr p 4 protein belongs to the alpha amylase family (InterPro no. IPR006046). Tyr p 4 protein has no transmembrane helices, and all amino acids of the Tyr p 4 protein are outside the membrane. ProtParam indicates that Tyr p 4 protein has a relative molecular weight of 58.4kDa and a theoretical isoelectric point (pI) of 6.76. The instability index (II) was computed to be 36.11, classifying the protein as stable. The grand average of hydropathicity is −0.431, indicating that Tyr p 4 protein is a hydrophilic protein. Tyr p 4 protein contains 42 phosphorylation sites, includes 27 serine sites (amino acids 28, 69, 95, 97, 166, 185, 193, 194, 221, 266, 361, 384, 424, 503, 504, and 505), five tyrosine sites (amino acids 196, 269, 362, 436, and 471), and one threonine site (amino acid 457). Secondary structure prediction shows that Tyr p 4 protein contains α-helices, β-sheets, and random coils. The analysis of the phylogenetic tree and sequence identity showed the closest genetic relationship between Tyr p 4 and Aca s 4 (Fig. 2 and Table 1).
Skin prick test and IgE-binding reactivity of recombinant Tyr p 4 proteinThe skin prick test of recombinant Tyr p 4 protein revealed seven + cases, twenty-two ++ cases, and one +++ case. The positive response rate was 19.2% (23 participants out of 120). The serum specific IgE test of recombinant Tyr p 4 protein revealed ten cases of Class 1, eight cases of Class 2, and two cases of Class 3. The total positive response rate was 16.7% (20/120). Sixteen participants were positive in both skin prick test and sera specific IgE-binding test, with a positive rate of 13.3% (16/120).
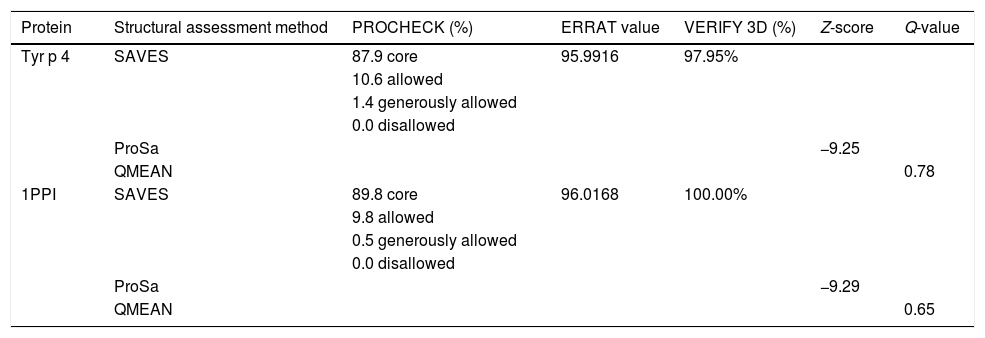
Homology modeling of Tyr p 4 protein tertiary structureThe sequence identity of alpha-amylase (PDB accession no. 1PPI) and Tyr p 4 protein is 52.32%. The parameters of the Tyr p 4 protein tertiary structure are similar to 1PPI (Table 2). PROCHECK shows high stereochemical quality of Tyr p 4 protein structure and the ERRAT value suggests high resolution of the tertiary structure of Tyr p 4 protein. VERIFY 3D suggests that the Tyr p 4 protein structure is favorable. The Z-scores of the template and Tyr p 4 protein show a high level of matching between the tertiary structures of the protein. The Q value suggests that the predicted model of Tyr p 4 protein is reliable. Based on this evaluation, we determined the tertiary structure of Tyr p 4 protein obtained by homologous modeling to be reliable. The tertiary structure of Tyr p 4 protein also contains α-helices, β-sheets, and random coils, and similar proportions of these elements are found in the secondary and tertiary structures. The proportions of α-helices, β-sheets and random coils in the secondary structure are 28.19% (15 domains), 18.34% (20 domains) and 53.47%, and are 25.87% (18 domains), 12.55% (13 domains) and 61.58% in the tertiary structure.
Evaluation of the tertiary structure of Tyr p 4 protein.
| Protein | Structural assessment method | PROCHECK (%) | ERRAT value | VERIFY 3D (%) | Z-score | Q-value |
|---|---|---|---|---|---|---|
| Tyr p 4 | SAVES | 87.9 core | 95.9916 | 97.95% | ||
| 10.6 allowed | ||||||
| 1.4 generously allowed | ||||||
| 0.0 disallowed | ||||||
| ProSa | −9.25 | |||||
| QMEAN | 0.78 | |||||
| 1PPI | SAVES | 89.8 core | 96.0168 | 100.00% | ||
| 9.8 allowed | ||||||
| 0.5 generously allowed | ||||||
| 0.0 disallowed | ||||||
| ProSa | −9.29 | |||||
| QMEAN | 0.65 |
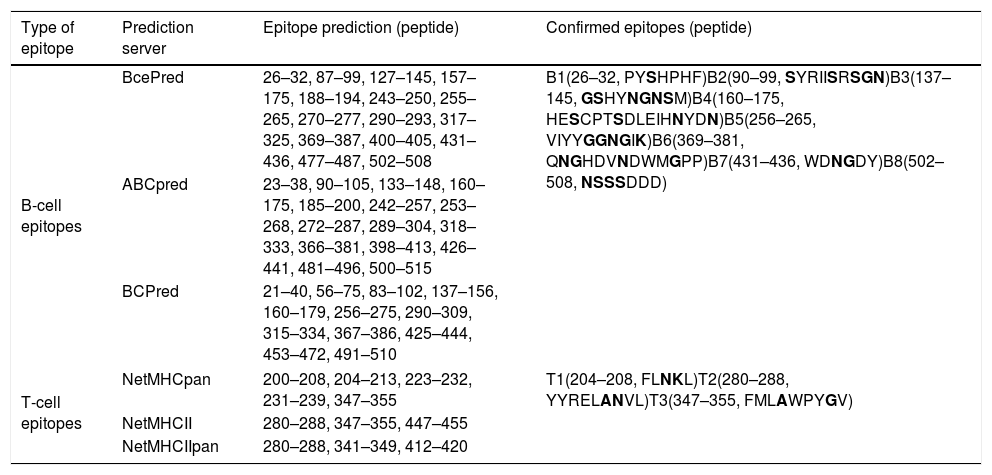
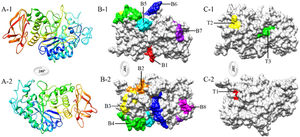
Eight B-cell epitopes (amino acid positions 26–32, 90–99, 137–145, 160–175, 256–265, 369–381, 431–436, and 502–508) and three T-cell epitopes (amino acid positions 204–208, 280–288, and 347–355) of Tyr p 4 were predicted (Fig. 3 and Table 3).
Predicted B and T cell epitopes of Tyr p 4.
| Type of epitope | Prediction server | Epitope prediction (peptide) | Confirmed epitopes (peptide) |
|---|---|---|---|
| B-cell epitopes | BcePred | 26–32, 87–99, 127–145, 157–175, 188–194, 243–250, 255–265, 270–277, 290–293, 317–325, 369–387, 400–405, 431–436, 477–487, 502–508 | B1(26–32, PYSHPHF)B2(90–99, SYRIISRSGN)B3(137–145, GSHYNGNSM)B4(160–175, HESCPTSDLEIHNYDN)B5(256–265, VIYYGGNGIK)B6(369–381, QNGHDVNDWMGPP)B7(431–436, WDNGDY)B8(502–508, NSSSDDD) |
| ABCpred | 23–38, 90–105, 133–148, 160–175, 185–200, 242–257, 253–268, 272–287, 289–304, 318–333, 366–381, 398–413, 426–441, 481–496, 500–515 | ||
| BCPred | 21–40, 56–75, 83–102, 137–156, 160–179, 256–275, 290–309, 315–334, 367–386, 425–444, 453–472, 491–510 | ||
| T-cell epitopes | NetMHCpan | 200–208, 204–213, 223–232, 231–239, 347–355 | T1(204–208, FLNKL)T2(280–288, YYRELANVL)T3(347–355, FMLAWPYGV) |
| NetMHCII | 280–288, 347–355, 447–455 | ||
| NetMHCIIpan | 280–288, 341–349, 412–420 | ||
Bold letters represent the hydrophobic amino acid residues.
Although it was a potential allergen, there is little information regarding the physical and chemical properties of Tyr p 4, its allergenicity, or antigenic epitopes. It has not been included in the official website of the systematic allergen nomenclature approved by the WHO/IUIS Allergen Nomenclature Database (http://www.allergen.org/). We determined the allergenicity of recombinant Tyr p 4 protein by skin prick test and ELISA. The skin prick positive rate was 19.2%, the rate of serum specific IgE positives was 16.7%, and the total positive response to both methods was 13.3%. The total positive rate of the skin prick test was higher than the serum specific IgE test, this is likely due to a false positive result on the skin prick test; thus, combining the two methods allows for a more accurate determination. Phylogenetic analysis shows that Tyr p 4 has the highest homology with Aca s 4 (Acarus siro) and Blo t 4 (Blomia tropicalis). Interestingly, all three are storage mites, and Blo t 4 induced positive skin prick tests in 10% of 200.34 Based on these results, the allergenicity of groups 4 allergens is weak compared to groups 1 and 2 allergens. Cross-reactivity sometimes occurs between the same group allergens of different mite species, but no reports on cross-reactivity between different mite species group 4 allergens have been found to date. This will be the focus of our further research.
Although Tyr p 4 protein contains α-helices, β-sheets, and random coils, the number of amino acids in these elements in the tertiary structure was found to vary marginally from the secondary structure. This discrepancy may be due to different structural prediction methods.
After determining the allergenicity of Tyr p 4, the B-cell and T-cell epitopes of Tyr p 4 were predicted. The properties of the antigen epitope, their number, and spatial configuration determine antigen specificity.35,36 α-helices and β-sheets have higher chemical bond energy and have difficulty forming epitope sequences. By contrast, β-turns and random coils are located in surface-exposed regions of a protein, which often contain epitope sequences.37 Epitopes usually contain 6–8 amino acids residues and in general contain <20 amino acid residues. In addition, allergen epitopes usually contain high proportions of hydrophobic amino acid residues, including Ala, Ser, Asn, Gly, and Lys.38 Based on the results of the epitope prediction software and the tertiary structure of the Tyr p 4 protein, the presence of eight B-cell epitopes and three T-cell epitopes is likely. However, these predicted epitopes require further experimental verification.
In conclusion, we demonstrate that the cloning, expression, characterization, allergenicity, and epitopes of recombinant Tyr p 4 protein has enabled an initial evaluation of its potency as an allergen in mite-allergic individuals. These findings provide a foundation from which to explore the structural biology and biochemistry of Tyr p 4 protein, thereby enabling future work with ASIT.
Conflict of interestThe authors have no conflict of interest to declare.
This work was financially supported by the National Sciences Foundation of China (NSFC 30060166, NSFC81001330, NSFC31272369, and NSFC31572319), the Jiangsu Overseas Visiting Scholar Program for University Prominent Young & Middle-aged Teachers and Presidents, the 333 project of Jiangsu Province in 2017 (BRA2017216), the Major Program of Wuxi Health and Family Planning Commission (Z201701), and the Primary Research & Development Plan of Jiangsu Province (Grant No. BE2018627).