Nowadays, cellular physiology is best understood by analysing their interacting molecular components. Proteins are the major components of the cells. Different proteins are organised in the form of functional clusters, pathways or networks. These molecules are ordered in clusters of receptor molecules of extracellular signals, transducers, sensors and biological response effectors. The identification of these intracellular signalling pathways in different cellular types has required a long journey of experimental work.
More than 300 intracellular signalling pathways have been identified in human cells. They participate in cell homeostasis processes for structural and functional maintenance. Some of them participate simultaneously or in a nearly consecutive progression to generate a cellular phenotypic change.
In this review, an analysis is performed on the main intracellular signalling pathways that take part in the cellular proliferation process, and the potential use of some components of these pathways as target for therapeutic interventionism are also underlined.
La fisiología celular es actualmente mejor entendida a partir de sus componentes moleculares interactuantes. Las proteínas son las principales biomoléculas que constituyen las células. Las diferentes proteínas se organizan en asociaciones formando conjuntos, vías, o redes funcionales. Estos conjuntos se encuentran organizados en moléculas receptoras de señales extracelulares, transductoras, sensoras y efectoras de respuestas biológicas. La identificación de las diferentes vías de señalización intracelulares en los diferentes tipos de células ha requerido largas jornadas de trabajo experimental.
Se han identificado más de 300 vías de señalización intracelular en las células humanas, las cuales participan en los procesos celulares básicos y especializados para mantener la homeostasis estructural y funcional. Varias de ellas participan de forma simultánea o cercanamente consecutiva en la generación de un cambio fenotípico celular.
En esta revisión analizamos las principales vías de señalización intracelular que participan en el proceso de proliferación celular y remarcamos la estrategia de utilizar algunos componentes de estas vías como blanco de un potencial intervencionismo terapéutico.
The capacity of human beings to see, perceive and respond to environmental signals depends on the activation of intracellular signalling pathways of their different constituting cells. These signalling pathways provide mechanisms to organise molecular information through cells, as it occurs with the central nervous system when it conducts the transduction of environmental information and organises the comprehensive response of the different body organs. In this way, sense organs are sensitive to certain kinds of stimuli (for example, light, pressure or sound waves), given that cell surface receptors bind to specific ligands. One cell may have dozens of different receptors that simultaneously receive signals and transmit them to the inside of the cell at the same time. All cells use specific intracellular signalling pathways to record incoming information, to translate it, to make different molecules interact and to produce a biological response, thus generating a specific phenotypic pattern.1
Signalling pathways mainly consist of a protein chain where proteins interact with each other in a previous sequence that was established through evolution. Plants and animals share certain basic intracellular signalling mechanisms; however, some organisational components are predominant, while others are less frequent and certain pathways are unique to each kingdom.
Over the last decades, a large number of intracellular signalling pathways, each with countless molecular components, have been identified. This characterisation has been conducted in the context of different cellular processes of specific types of cells, in conditions of both health and disease. A general understanding has been reached in relation to how these groups of molecules work in terms of circuits and complex systems.
The normal development and functioning of multicellular prokaryotes and eukaryotes depend on a consecutive series of cellular interactions based on biochemical changes. The different cellular stages are mainly due to genetic transcription patterns that, upon translation, produce the interaction with other molecules and, finally, regulate different cellular functions.
Cloning and sequencing of DNA segments and purification and sequencing of proteins have led, for example, to understand how some oncogenes code for growth factors (such as v-Sis) and to recognise their receptors (such as ErbB) and signalling pathway proteins (such as v-Src). Likewise, it has been demonstrated that the transduction of signals occurs through both second messengers, and protein–protein interactions and protein–nucleic acid interactions, or how epigenetic cellular conditions modify the effector response due to extracellular signals.2
In this article, the general principles of intracellular signalling pathway functioning will be described, and the modular organisation of pathways participating in the cellular proliferation process will be analysed, as an example of a complex molecular model that cells use to conduct various cellular processes.
Proteins as participating biomoleculesProteins are the most common and complex biomolecules within cells, and they constitute their structural and functional elements. Proteins are functional molecules of biological work. Based on their direct genetic coding in human cells, there would be 22,000 different proteins; however, this number is increased more than 10 times due to the different transcription combinations of genes and post-transductional changes that occur in the endoplasmic reticulum and Golgi apparatus. They are essentially composed of 20 types of amino acids in different proportions. There are small proteins (40–80 amino acids), medium and large (more than 10,000 amino acids), and they are biochemically organised into amino acid chains (primary structure), into helices and folded sheets (secondary structure), into small three-dimensional molecules formed by attraction and repulsion between amino acids (tertiary structure), and into large three-dimensional molecules formed by the joining of smaller structures (quaternary structure). Several protein affinity and sequencing studies conducted over the last decades have demonstrated that specific peptide segments or intramolecular domains work in a semi-independent manner and are commonly identified with specific functions, such as kinase and phospholipid binding domains. Proteins are biomolecules that interact with other kinds of small molecules or with adjacent proteins and modify them by producing new intramolecular changes, which are biological activation indicators. Two main protein function activation or repression mechanisms are phosphorylation/dephosphorylation and GTP or GDP-binding.1,3,4
Various kinds of proteins, due to their wide range of type, shape, domain, response and interconnectivity with other proteins, are usually main components in the reception, transduction, regulation and control of extracellular and intracellular signals. The different kinds of bioactive proteins are essential for the biochemical response in intracellular signalling pathways, which ultimately lead to a structural and functional phenotypic change in several cells. Each live cell contains molecular participation mechanisms that “turn on” and “turn off” the responses for different electrical and chemical signals.
Cellular signalling systemsTransduction of signals, sensors and molecular effectorsIntracellular proteins are organised into groups by function, and these groups are generally located in specific subcellular compartments. The basic components of an intracellular signalling pathway comprise the following molecules: the extracellular signal, signal receptors, transducers, sensors, effectors and, finally, the activation of the cellular response to express a specific phenotype.1,5 Extracellular signals may be long-distance or short-distance, are generally large and hydrophilic molecules, and correspond to neurotransmitters, hormones (including local hormones), cytokines, growth factors, cell surface molecules and sensory stimulation molecules. These signals are bound to receptors by means of 4 mechanisms: juxtacrine, autocrine, paracrine and endocrine. Extracellular signals may be free in extracellular fluid or embedded in the extracellular matrix, and the response is dependent on whether the specific receptor is present in the cell. Receptor proteins detect the signal at the cell membrane and transmit it to internal transducer molecules, which consist of mainly 3 types: receptors associated with ion channels (they act in the simplest and most direct manner), receptors associated with G-proteins (monomeric and heterotrimeric) and receptors associated with enzymes (mostly with kinase activity) that help to control complex cellular behaviours.3,4 Many extracellular signalling molecules interact with more than one kind of receptor. Signals reaching receptors associated with G-proteins or enzymes are transferred to complex transmission systems formed by series of intracellular signalling molecules and their activation produces specific covalent and conformational changes that turn them into substrates of the following reaction within the signalling pathway. Except for some small molecules, such as cyclic GMP, cyclic AMP and Ca2+, most signalling molecules are proteins. In general, small molecules considerably amplify signals and act as a stimulus for the simultaneous activation of several surrounding signalling pathways. Activated proteins generally go through one or few pathways, and some of them act as a sequential part of the pathway, others act as molecular hubs for one or several pathways, and others act as scaffolding molecules.1,6 Transducer molecules pass on the signal to intracellular sensor molecules and effector molecules; the first may be intermediaries or final elements of specific signalling pathways; some effector proteins (for example, transcription factors) may act on single or multiple molecular targets and conduct processes, such as exocytosis, phagocytosis, actin remodelling, metabolic pathway activation and genetic expression. De novo protein synthesis is generally required for the implementation of molecular interaction programmes. The different kinds of signalling pathways used by cells to regulate their different interrelated cellular processes are interconnected through hub molecules, forming complex networks with several signalling pathways. During their activation, signals may travel through some signalling pathways in particular, using a preferred or canonical route, or eventually using an alternative or non-canonical route.1 The termination of the activation of intracellular signalling pathways implies stopping the local process in one or several components by means of the action of opposing enzymes (for example, kinase vs. phosphatase) or specific inhibitors. From an evolutionary point of view, the genome contains all the structural codes for the components of the different signalling pathways. Thus, for example, in the differentiation process, each specialised cell deactivates the coding for molecules participating in the cellular proliferation process and activates the coding for molecules participating in specialised catabolism processes or specialised functioning processes for structural and functional phenotype programmes implemented by terminally differentiated cells.
The activation of intracellular signalling pathways is carefully organised in time and space. The signalling process starts outside the cell and continues inside it, while cellular response occurs in an inside-out fashion. For each “on” molecular activation mechanism, there is an “off” deactivation mechanism. The activation language of the pathway is established by direct protein–protein interaction or through its covalent modifications, such as phosphorylation and acetylation, caused in the pathway by the upstream biomolecule. This activation requires the adequate temporospatial coordination of its components. Some cellular response patterns, such as growth increase and cell division, imply changes in both genetic expression and synthesis of new proteins (slow responses), while changes in cellular movement, secretion or metabolism do not require the participation of nuclear machinery and occur more rapidly.
The “on” and “off” functioning in intracellular signalling pathways has a high level of cellular plasticity, and these signalling pathways are constantly being remodelled by external or internal signals required for the maintenance of cellular homeostasis. Mechanisms that deactivate signals are as important as those which activate them. Complex cellular behaviours are usually controlled by networks of protein-kinases; these kinases can also modulate components of their own signalling pathways, components of other surrounding signalling pathways or of related pathways through hub molecules. The adequate integration of multiple intracellular signalling pathways regulates complex processes, such as development, proliferation, differentiation, response to stress, apoptosis, etc. Alterations in the regulation of signalling pathways lead to the inadequate activation of the cellular response and to many diseases. The investigation of intracellular signalling pathways in different types of cells is an active area and there are constantly new discoveries.1,6,7
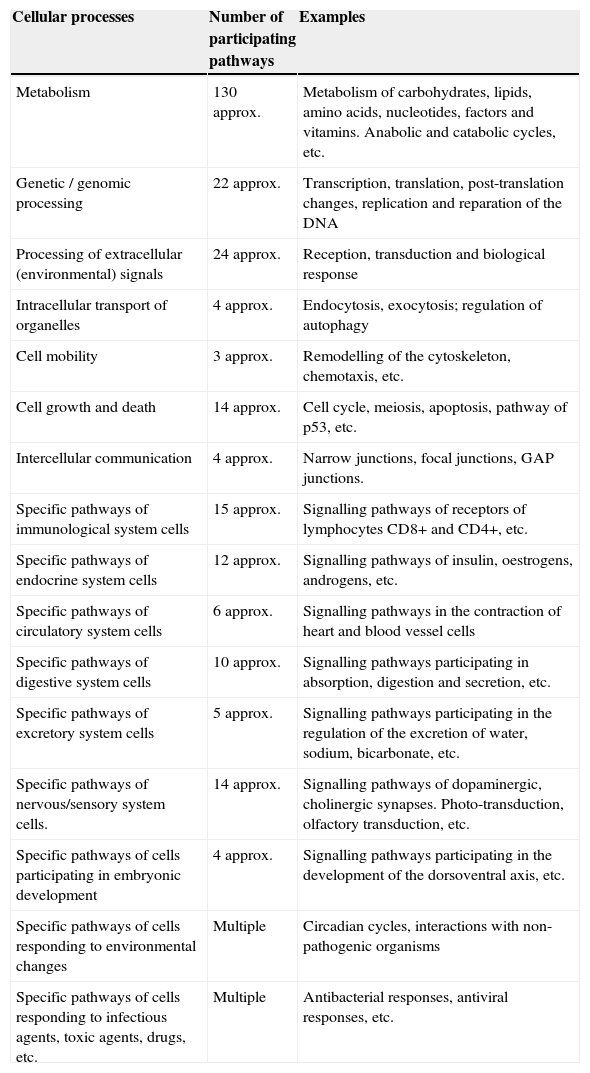
Main intracellular signalling pathways participating in various structural and functional cellular processesThe biomolecular pathways of intracellular signalling are groups of proteins and other biomolecules arranged in cascades of spatio-temporal interactions and they are responsible for cellular phenotypic characteristics. Each molecular signalling pathway is generally located in a separate subcellular compartment.6 Information on signalling pathways or signalosome is essential for the construction of models of biological systems. The Kyoto Encyclopaedia of Genes and Genomes (KEGG) is one of the databases that contains the main intracellular signalling pathways participating in the regulation of the main cellular processes (Table 1)8; some of them use small messenger molecules, also known as second messengers, for the transmission and amplification of messages, while others mainly use proteins as messengers and internal sensors.1 The first group is formed, for example, by the pathways of cyclic AMP, cyclic/NAADP ADP-ribose, voltage-operated channels (VOC), receptor-operated channels (ROC, such as K+, Ca2+ or Cl−), the activation pathway of phospholipase-C (PLC) that breaks down phosphatidylinositols of the cell membrane, the activation pathway of phosphatidylinositol-3-kinase (PI3K, which phosphorylates PIP2 and forms PIP3), the nitric oxide/cyclic GMP pathway, the redox signalling pathway, the phospholipase-D (PLD) pathway, the sphingomyelin pathway, the AMP pathway, the NAD pathway and many other pathways participating in the metabolism. The second group is formed by mitogen-activated kinases (MAPK) pathways, the nuclear factor B (NF-B) pathway, the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway, the SMAD, WNT, Hedgehog, Hippo, Notch pathways, the endoplasmic reticulum stress pathway and several other pathways.
Number of intracellular signalling pathways participating in the main structural and functional cellular processes.
| Cellular processes | Number of participating pathways | Examples |
|---|---|---|
| Metabolism | 130 approx. | Metabolism of carbohydrates, lipids, amino acids, nucleotides, factors and vitamins. Anabolic and catabolic cycles, etc. |
| Genetic / genomic processing | 22 approx. | Transcription, translation, post-translation changes, replication and reparation of the DNA |
| Processing of extracellular (environmental) signals | 24 approx. | Reception, transduction and biological response |
| Intracellular transport of organelles | 4 approx. | Endocytosis, exocytosis; regulation of autophagy |
| Cell mobility | 3 approx. | Remodelling of the cytoskeleton, chemotaxis, etc. |
| Cell growth and death | 14 approx. | Cell cycle, meiosis, apoptosis, pathway of p53, etc. |
| Intercellular communication | 4 approx. | Narrow junctions, focal junctions, GAP junctions. |
| Specific pathways of immunological system cells | 15 approx. | Signalling pathways of receptors of lymphocytes CD8+ and CD4+, etc. |
| Specific pathways of endocrine system cells | 12 approx. | Signalling pathways of insulin, oestrogens, androgens, etc. |
| Specific pathways of circulatory system cells | 6 approx. | Signalling pathways in the contraction of heart and blood vessel cells |
| Specific pathways of digestive system cells | 10 approx. | Signalling pathways participating in absorption, digestion and secretion, etc. |
| Specific pathways of excretory system cells | 5 approx. | Signalling pathways participating in the regulation of the excretion of water, sodium, bicarbonate, etc. |
| Specific pathways of nervous/sensory system cells. | 14 approx. | Signalling pathways of dopaminergic, cholinergic synapses. Photo-transduction, olfactory transduction, etc. |
| Specific pathways of cells participating in embryonic development | 4 approx. | Signalling pathways participating in the development of the dorsoventral axis, etc. |
| Specific pathways of cells responding to environmental changes | Multiple | Circadian cycles, interactions with non-pathogenic organisms |
| Specific pathways of cells responding to infectious agents, toxic agents, drugs, etc. | Multiple | Antibacterial responses, antiviral responses, etc. |
approx.: approximately.
The elucidation of intracellular signalling pathways has led to the conduction of several trials with protein genomic and biochemical techniques in dynamic models of normal and tumour cells.7 The identification of a specific pathway takes years of experimental work. The participation and importance of a specific protein in the signalling process is determined using different experimental strategies which are frequently combined, such as the introduction or deactivation of a gene that codes for said protein, the assessment of its expression and its post-transductional activation/deactivation, all of which is related to a specific phenotypic cellular effect. The deactivation of proteins participating in intracellular signalling pathways has been studied using genetic molecular techniques aimed at blocking or reducing their genetic expression, such as knock-out and knock-down techniques.9 To explore proteins activated by phosphorylation, specific antibodies are used or proteins marked with radioactive ATP. To identify interacting proteins, co-immunoprecipitation assays are conducted and, upon identification of interactive proteins, their active domains can be recognised using recombinant DNA assays (targeted mutagenesis) through the creation of mutant proteins, and their interaction and importance can be analysed. A strategy that facilitates the identification of proteins participating in any specific intracellular signalling pathway is to conduct a populational study of tens of thousands of fruit flies or nematodes treated with a mutagen, and to compare these to the control group, so as to identify mutants with an altered phenotypic trait due to the inappropriate functioning of one or several signalling pathways. In these cases, altered genes that code for proteins participating in signalling cascades are identified and compared to their corresponding control groups.1,4 One of the cases that requires the greatest amount of experimental work is to determine the location of a participating protein either downstream or upstream a signalling pathway, compared to the other participating proteins.
The cumulative experimental study in this area has identified a great number of intracellular molecular signalling pathways related to: global cellular functions, such as metabolism, proliferation, apoptosis; specialised cellular functions, such as immunological response, transcription factor interactions, and protein-protein interactions; and cellular functions altered in the presence of different diseases.10 To analyse this large amount of information, it was necessary to resort to bioinformatics for the construction of registration databases and molecular interactions (nodes), such as BioPAX, Pathway Interaction Database, Reactome, BioCarta, KEGG, GenMAPP, Genenetwork, Cytoscape, NetPath, among others. Many of these were constructed with the collaboration of interdisciplinary groups, for instance the NetPath database, which is a project created by both the Pandey Lab (John Hopkins University) and the Institute of Bioinformatics (Bangalore, India). Some mega databases, such as that of the National Centre of Biotechnology Information (NCBI), Pathguide and WikiPathways11,12 form multiple databases of modules or nodes related to cellular signalling pathways from various types of normal cells and several diseased cells. The KEGG,13 based on the integration of genomic, transcriptomic, proteomic and metabolic data, has organised the maps corresponding to more than 300 human intracellular signalling pathways related to metabolism, the processing of genetic information, the processing of responses to environmental changes, basic cellular processes, different types of differentiated cells from body organs and systems and particular models of different human diseases,8 thus demonstrating that each specialised or basic cellular process requires the participation of some or multiple intracellular signalling pathways that work in a simultaneous manner. These interacting groups of intracellular signalling pathways that generate a phenotypic change are called modules.7 Basic cellular process pathway modules include molecular transport and catabolism, cellular mortality, cellular growth, death and communication (all of which have specific sub-pathways). Pathway modules identified with diseases include those related to some cancers (colorectal, pancreatic), immunological diseases (asthma, erythematosus lupus), neurodegenerative diseases (Alzheimer's, amyotrophic lateral sclerosis), addictions (to cocaine, to morphine), cardiovascular diseases (cardiomyopathic, hypertrophic), endocrine and metabolic diseases (diabetes mellitus types 1 and 2), infectious diseases (infection by salmonella, influenza A).1,6 These maps make it possible to understand how the overactivation or repression of gene expression resulting from mutations, epimutations or changes in the number of copies in a specific region of the human genome modify the activation or repression of specific intracellular signalling and lead to structural and functional changes in cells. In the same way, the study of these molecular maps facilitates possible interventions to use or design molecules with biochemical affinity and target altered molecules, so as to block or modify the specific biochemical reaction, produce a thermodynamic change in the intracellular signalling pathway and, finally, generate a change in the genetic expression and cell phenotype. The correction of the functioning in altered intracellular signalling pathways may limit or reverse cellular and tissue damage and recover the homeostasis.14 As an example of the participation in several intracellular signalling pathways in some modules or programmes of cellular phenotypic processes, the main intracellular signalling pathways participating in the division and proliferation of eukaryotes will be briefly analysed.
Module of intracellular signalling pathways participating in cell division and proliferation cyclesPotential targets of molecular interventionsEukaryotes, depending on the activation/deactivation, deviation or modulation in the networks of signalling pathways of the cell division and proliferation cycle, may pass through 4 different stages: senescence, apoptosis, differentiation and proliferation.1,3
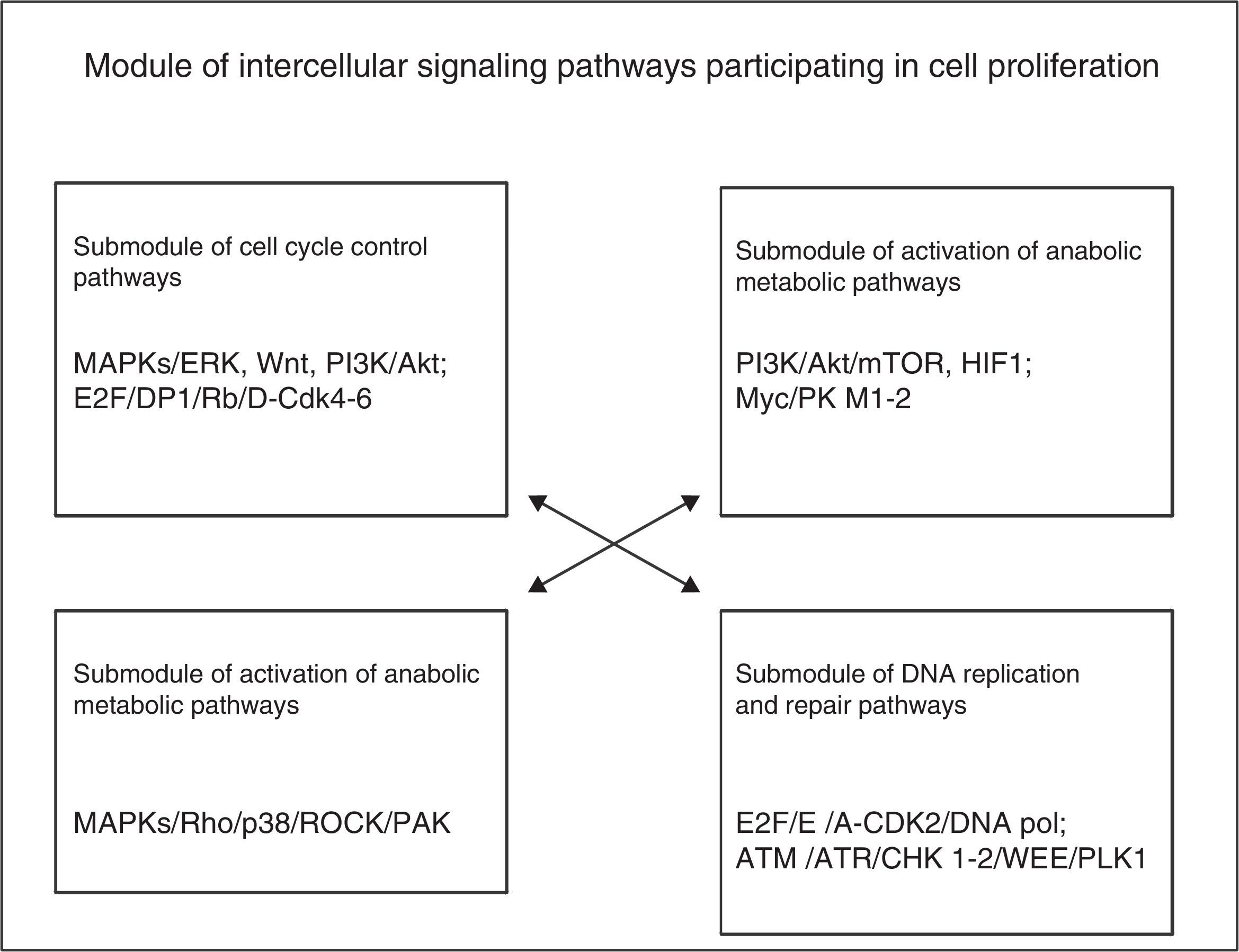
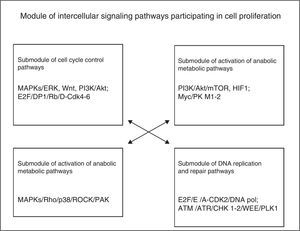
The module of cell division and proliferation consists of 4 main groups of intracellular signalling pathways: those for the control of the cell cycle, those for the activation of metabolic-anabolic pathways, those for activation or remodelling of the cytoskeleton, and DNA replication and repair. In the cell proliferation process, cells self-reproduce by means of intracellular growth and division in 2 identical copies, which implies that the cell needs, at least, to increase its biomass and replicate its genome. Different growth factors (GF) activate cells during interphase so that they develop during their cell cycle and increase their cell mass by increasing the biosynthesis of macromolecules through the activation of anabolic signalling pathways. GF activate the MAPK/Ras/RAf/ERK pathway to produce the synthesis of 3 types of cyclins D (D1, D2 and D3) and other positive mitosis-regulating proteins (Myc and Jun); cyclin D1 binds to CDK4/6 to form an effective complex to activate several substrates required to initiate the cell cycle, such as Myc and AP-1. Cyclin D is regulated by the MAPK/Ras, β-catenin-Tcf/LEF, PI3K and Rho/FAK. The availability of cyclin D in cells is controlled by a balance between its Ras/Raf/MAPK-dependent synthesis and its Ras/PI3K/AKT, GSK3 and SCF-dependent synthesis.1
The cell cycle is divided into 2 main phases: phase M and interphase. Phase M includes the consecutive phenomena of mitosis and cytokinesis. Interphase is divided into phases G1, S and G2, phase S being equivalent to the period of DNA synthesis. In phase G1, the cell increases its size and activates the replication of DNA; in phase G2, the cell continues to grow and mitosis is activated; in phase M, the cell stops growing and is divided into 2 daughter cells by means of cytokinesis. After cell division, or “post-mitosis” phase, each daughter cell goes through the interphase and is prepared for 2 situations: another cell division or a non-proliferation status. The following paragraphs summarily describe the main molecules participating in the signalling pathways of the cell division and proliferation/growth cycle (for more details, refer to Berridge, 2012).1,15
Phase G1 is called growth phase, during which millions of proteins, enzymes, biolipids and nucleotides are formed. These are required for the biosynthesis of the cytoplasm, cell membranes, organelles, DNA and RNA. The duration of G1 varies greatly from one cell to the other and is under the control of protein p53. Intracellular signalling pathways participating through GF are mitogen-activated protein kinases (MAPK) pathways or cascades, the WNT pathway that acts by means of β-catenin and the PI3K pathway. These increase transcription in cyclin-dependant cyclin/kinase (CDK) complexes and, in particular, in cyclin D, which acts on the retinoblastoma protein to remove its inhibitory effect towards E2F, which activates the formation of cyclins E and A, and these cause cells to enter into phase M. In contrast with these positive effects, pathways of transformation growth factors (TGF)/Smads act by preventing the cell from starting the cell cycle through the expression of CDK inhibitors (INK4 and Cip/Kip families) and p53. In particular, p53 stops the cycle when DNA is not correctly replicated, thus producing proteins p21, GADD45 and 14-3-3, and promoting the transcription of apoptotic factors.15
As mentioned before, after the cell receives GF, cyclin D is rapidly synthesised, and this continues to occur throughout the cell division cycle. However, cyclin E appears as a peak towards the end of G1; cyclin A only appears in phases S and G2; and cyclin B is expressed in phases G2 and M, while CDK are constitutively produced. In contrast with these cyclin increases, the cyclin/CDK inhibitor is reduced as from the beginning of phase G1, which continues to occur throughout all the phases of the cell cycle. Cyclins/CDK work as serine-threonine kinases: cyclin D/CDK4-6 phosphorylates protein Rb, so the family of transcription factors E2F activate the transcription of the genes of cyclins E and A, of phosphatase cdc25A that remodel the chromatin. Then, cyclin E/CDK2 and cyclin A/CDK2 are activated to start with the DNA synthesis, while the function of cdc25A allows for the activation of cyclin B/CDK1 to begin the mitotic spindle assembly.16
MAPK pathways are generally the most important signalling pathways that regulate cell proliferation, and cyclin D1 is the main sensor of extracellular signals that drives the progression phase of the cycle from G1 to S.17 The complexity of these pathways and that of the cyclin D1 expression will be briefly analysed below.
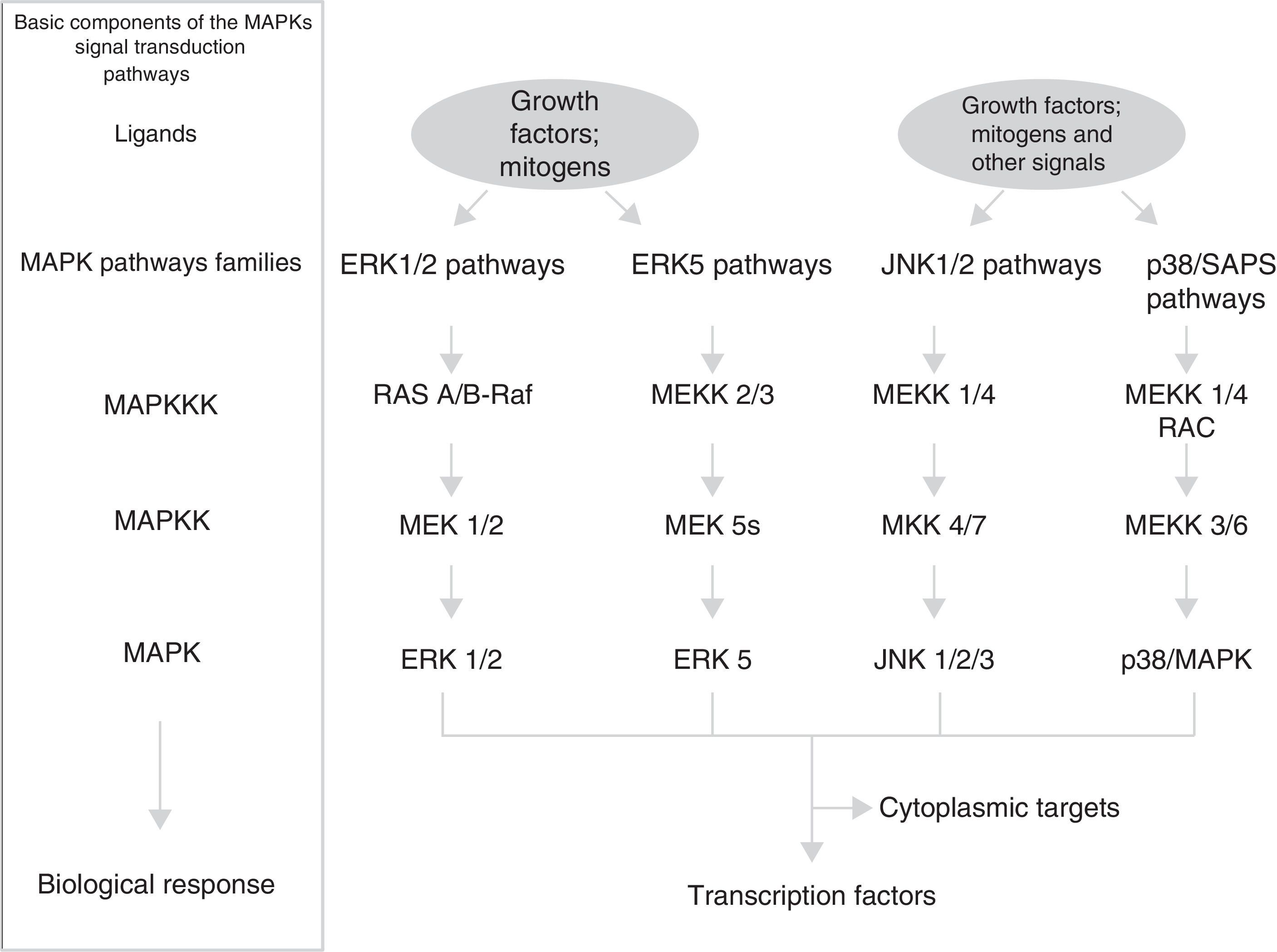
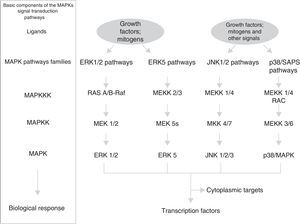
MAPK pathwaysThese pathways participate not only in the regulation of cell proliferation, but also in the regulation of cell differentiation and death. The module of MAPK pathways contains from 3 to 5 components or rows (horizontal) of protein-kinases (Fig. 1): a MAP-kinase-kinase-kinase (MAPKKK) that, at the same time, consecutively activates a MAP-kinase-kinase (MAPKK) and a MAP-kinase (MAPK). This phosphorylates serines and threonines of proteins that lead to changes in their function and in gene expression. The MAP-kinase group has 3 families of interrelated components (vertical distribution) that receive, transmit and respond to different extracellular signals: ERK, JNK and p/38/SAPK; in particular, the family of signalling pathways regulated by extracellular signals (ERK) is divided into 2 sub-families (Fig. 1): small ERK1 and ERK2, and large ERK5.17 Each cascade or family/sub-family responds to different growth factors, differentiation factors, stress extracellular signals and inflammatory cytokines; they contain different scaffolding proteins and activate different proteins or gene expression (some of which are similar). This means that MAPK pathways jointly correspond to a system of hundreds of proteins (many of them correspond to isoforms produced by the processing of MAPK genetic expression) which regulate different cellular physiological functions by acting as transcription factors or activating nuclear/cytoplasmic enzymes.18,19 The main temporospatial mechanisms proposed so that MAPK pathways regulate cellular functions are the following: (a) different duration and intensity of extracellular signals; (b) different interactions between components of the 3 families and different scaffolding proteins; (c) different interactions between the 3 MAPK families; (d) different subcellular compartmentalisation; and (e) intrinsic complexity of the vertical distribution components of each cascade.17
Cyclin D1Cyclin D1 is the sensor that integrates extracellular signals with molecular machinery that, bound to CDK4-6, activates the progression of the cell cycle. Different transcription factors, such as AP-1, SP-1, E2F, OCT-1, are induced by the activation of pathways ERK1/ERK2 and ERK5, or pathways PI3K/Akt and Wnt/β-catenin (or others, such as NF-κB, JAK/STAT), thus promoting the expression of cyclin D1 by binding to their promoter. Some groups of transcription factors organised in functional complexes, such as MuvB, B-Myb, FoxM1, DREAM, E2F, use different feedback circuits to interact at different times with cyclin D1 and lead to the progression of the cell cycle in its initial stages and in the final stages of phase G1.20,21
As previously explained, proliferating cells significantly increase ATP production and the synthesis of biomolecules, including lipids, proteins and nucleic acids, so they re-programme their metabolic pathways and, instead of using catabolic cycles, such as that of mitochondrial oxidative phosphorylation, they start to use anabolic cycles, such as the aerobic glycolysis pathway and other anabolic routes.22,23 Glycolysis produces ATP more rapidly due to the overexpression of several enzymes used in this pathway and provides a greater number of intermediary molecules for the biosynthesis of macromolecules. The regulating axis of the cell cycle CDK4/pRb/E2F1 is also an important factor in the control of the energetic status and the biosynthetic anabolic process.23
Upon activating the pathway PI3K/Akt, the cell increases the use of nutrients, particularly glucose and glutamine, and the use of carbons is re-directed by glycolytic enzymes, such as pyruvate kinase, towards the synthesis of macromolecules. During this phase, GF are bound to tyrosine-kinase receptors and other receptors associated with proteins G, which stimulate the formation of inositol 1,4,5-trisphosphate (IP3) and diacylglycerol, which release Ca2+ and turn on biosynthesis pathways. The mTOR kinase, a serine/threonine kinase inhibited by rapamycin, and the transcription factor Myc through the pathway PKB/TSC1/2 are the molecules that coordinate the synthesis of proteins and nucleic acids, respectively. Signalling pathways of cell cycle progression and metabolic pathways have a two-way relation, since they both execute feedback controls between one another.24
Apart from these 2 groups of signalling pathways, another group of signalling pathways participates in cell cycle progression: those related to intracellular cytoskeleton remodelling, which lead to changes in the shape of the cell, chromosome alignment, organelle movement, and polarisation and separation of the 2 daughter cells at the end of mitosis; in these, Rho-GTPase proteins work as hubs. Rho proteins, members of the Ras superfamily, regulate the assembly and organisation of cytoskeleton formations composed of actin and microtubules, which contribute to the formation of the mitotic spindle and its binding to the kinetochore, coordinate the contractile ring and cell separation during cytokinesis, etc. Rho proteins include RhoA, Rac1 and Cdc42, and are interconnected with the signalling pathways MAPK, Wnt/β-catenin and PI3K; in particular, they participate in the JNK and p38/SAPK cascades of MAPK pathways, in which Ras interact directly with each of the 3 proteins,18 transmitting the signal downstream through the rows of protein-kinases, activating cytoskeleton remodelling proteins, such as ROCK, MKLP1, PKN and mDia.25,26
In phase S, the genome, chromosomes and histones continue to replicate, thus achieving full duplication. The cyclin E/CDK2 complex contributes to cell cycle progression by phosphorylating Rb, removing p21, p27 and p57, and starting DNA synthesis. Cyclin/CDK complexes recruit and refine the molecular machinery of DNA replication and repair. DNA synthesis goes through 2 large signalling pathways: synthesis itself and repair of previous DNA damage caused by exogenous or endogenous agents (for example, oxygen reactive compounds)27 or the correction of errors that occur during the replication process (pathways for the repair of non-complementary base pairing). DNA damage response (DDR) pathways are divided into pathways that repair the damage of one chain (nucleotide and base excision repair) or pathways that repair the damage of the 2 chains (homologous recombination and pairing of non-homologous terminal segments), using molecules that recognise the damage, such as Ku70/80, RAD50, PARP, FANC, BRCA; transducer molecules, such as ATM and ATR; and effector molecules, such as CHK1 and CH2.28,29 P53 works as one of the main coordinating molecules between cell cycle pathways and DNA synthesis pathways.
In phase G2, the cell continues with the biosynthetic metabolic phase, verifies the fidelity of the replicated DNA and completes the formation of the mitotic spindle. The cyclin B/CDK1 complex controls the 2 final phases of the cell cycle.
In phase M, the nucleus is ruptured and divided during the sub-phases prophase, metaphase, anaphase, telophase, and the process is completed during the cytokinesis. In phase M, the nucleus, cytoplasm, organelles and cell membrane are divided into 2 cells that contain identical amounts. The main events of this phase are rupture of the nuclear envelope, assembly of the mitotic spindle, separation of chromosomes and cytokinesis. Cyclin B/CDK1 leads to the formation of the mitotic spindle, which organises the polymerisation of microtubule networks to form the organising centre (microtubule-organising centre, MTOC) and regulate its orientation; in this case, the mitotic spindle assembly signalling pathway participates in the anaphase activation complex (anaphase-promoting complex, APC).1,5 Chromosome separation is regulated by the APC/Plk1/SCF complex which hydrolyses cohesin molecules. During cytokinesis, the MgcRacGAP/RhoA complex restructures cytoskeletal filaments to form an actinomyosin contractile ring.26
According to the above description, the phenotypic module used for cell progression during the cell division cycle and cell proliferation requires the participation of 4 submodules of intracellular signalling pathways in a simultaneous or nearly consecutive manner, as shown in Fig. 2. Each pathway may contain more than 200 components organised in horizontal rows (upstream and downstream arrangements) and vertical rows (different cascades or families), as it occurs with MAPK pathways in particular. Furthermore, each signalling pathway submodule is interconnected, either directly or indirectly, with other pathway submodules related, either directly or indirectly, to the phenotypic effect of cell proliferation.
The analysis of this module leads to the conclusion that the generation of cellular phenotypic traits or characteristics probably involves some or multiple predominant intracellular signalling pathways and multiple subpredominant signalling pathways that are coordinated in a simultaneous or nearly consecutive manner. These involve a large number of molecular components that may potentially be subject to targeted therapeutic interventions; these molecular targets may mainly correspond to hubs of signalling cascades of only one pathway or to hubs regulating the interconnection with other essential pathways to achieve a specific cellular phenotypic change.
In this way, in cell proliferation, different groups of researchers began a large number of preclinical and clinical, experimental, phase I–II trials to try to modify cell cycle reprogramming in cancer cells through the pharmacological administration of active agents aimed at altering hubs of their predominant and subpredominant signalling pathways. One of these strategies involved the use of some flavonoids and purine-derived compounds that regulate different CDK through their inhibitory effect on serine-threonine kinase domains and, thus, their high proliferation rate.30
The identification of alterations in the regulation of MAPK signalling pathways is an essential biomarker in the oncogenesis of different tumours. For instance, in the case of thyroid papillary carcinoma, 3 parts of MAPK pathways, the receptor and the transducer molecules Ras and BRAF, are involved in more than 70% of the cases, and their differential association with the clinical behaviour of the disease has been observed in some patients. In patients with thyroid papillary carcinoma where conventional treatment is ineffective, the use of tyrosine-kinase receptor inhibitors is a therapeutic alternative.31 The determination of alterations in the components of this pathway through the identification of structural mutations and changes in the expression patterns of this receptor or RAS/BRAF in tumour cells are potential therapeutic targets.32,33
ConclusionsThe identification of intracellular signalling pathways participating in the generation of different basic and specialised processes that occur within cells allows for the understanding of underlying molecular mechanisms of cell physiology. The elucidation of intracellular signalling pathways takes years of experimental work.
For the generation of different cellular phenotypic changes, cells frequently use some or multiple simultaneous or nearly consecutive intracellular signalling pathways, as observed in the cell proliferation process. The identification of the main molecules that regulate signalling pathways in pathological cellular processes makes it possible to try to modify and reverse or limit the alterations in basic and specialised cellular processes and, therefore, alterations in tissues or organs. A more rational era of molecular therapy has begun.
Conflict of interestThe authors declare that there are no conflicts of interest.
Please cite this article as: Valdespino-Gómez VM, Valdespino-Castillo PM, Valdespino-Castillo VE. Interacción de las vías de señalización intracelulares participantes en la proliferación celular: potencial blanco de intervencionismo terapéutico. Cir Cir. 2015; 83: 165–174.