There are nanoparticles with remarkable antibacterial characteristics and aptamers able to recognize specific pathogenic bacteria with high affinity and specificity. The combination of both systems has been used to design rapid bacterial detection methods with excellent detection limits. Likewise, the synergism between aptamers and nanoparticles have allowed to optimize the antimicrobial activity of antibiotics and other nanostructures providing them with activity bacterium-specific, turning into attractive and promising tools to fight against bacteria resistant to multiple antimicrobials.
Existen nanopartículas con características antibacterianas destacables y aptámeros capaces de reconocer con gran afinidad y especificidad a determinadas bacterias patógenas. La combinación de ambos sistemas se ha utilizado en el diseño de métodos rápidos de detección bacteriana con excelentes límites de detección. Asimismo, el sinergismo entre aptámeros y nanopartículas ha permitido optimizar la actividad antimicrobiana de antibióticos y otras nanoestructuras dotándolos de actividad bacteria-específica, convirtiéndolas en herramientas atractivas y prometedoras frente a las bacterias resistentes a múltiples antimicrobianos.
Morbidity and mortality rates for bacterial infections have increased due to the increased frequency of multidrug-resistant (MDR) bacteria,1,2 which has reduced the efficacy of available eradication therapies3 and established a major health problem with serious economic and social consequences.4,5
This “antibiotic resistance crisis”6 has mainly been generated by the extensive and inappropriate use of antibiotics,4 but also by conventional diagnosis methods involving culture and biochemical tests (standard method), which take several days to identify infectious agents, allowing the infection to progress. It is therefore necessary to develop rapid methods for bacterial diagnosis,5 to design therapies that evade bacterial resistance mechanisms3,6 and/or to improve the action of the existing antibiotics, where nanotechnology appears to be a promising tool.4
AptamersAptamers are short, single-stranded nucleic acids, selected in vitro by a process known as systematic evolution of ligands by exponential enrichment (SELEX) to recognise a range of specific targets.7,8
Aptamer-target recognition is achieved through structural compatibility and combining of various non-covalent interactions,9,10 establishing dissociation constants usually in the picomolar to nanomolar range for targets with high molecular weight, and nanomolar to micromolar range for targets with low molecular weight.9,11 Aptamers are able to discriminate between enantiomers and molecules that structurally differ in only one functional group.12 Their small size and low molecular weight,11in vitro synthesis,7,12 stability under a broad range of conditions8,9,11,12 and lack of or low toxicity in vivo,9 have also made them attractive molecules for the development of new diagnostic and therapeutic strategies for infectious agents.8,9,11,12
Additionally, these biomolecules can be easily modified to improve their biostability against nucleases,8,9,11,12 increase their bioavailability, pharmacokinetic properties and affinity, evade the immune response,9 and even be coupled to reporter molecules, functional groups or nanoparticles (NP) to increase their applicability.7
NanoparticlesNPs are a range of small materials measuring less than 100 nm, the properties of which depend on their size, shape, distribution and chemical formulation.1,2,13 Their high surface area to volume ratio gives them a high degree of reactivity and unique interactions with biological systems.1 Moreover, the localised surface plasmon resonance (LSPR) commonly manifested by metal NPs under photonic or electromagnetic stimuli gives them optical properties equivalent to 10 fluorophores,13 so they could be used for the development of diagnostic tools.13
There are also NPs which, at non-toxic doses to human cells, have outstanding antimicrobial qualities against various pathogens and their MDR variants,1,13 where metal NPs appear to be promising,1 as they produce reactive oxygen species, constantly release metal cations1,3,14 and are generally positively charged, contributing to their adhesion and accumulation in the bacterial outer membrane,3,14 dissociating it and causing the elimination of the proton gradient and the subsequent exit of the cytoplasmic content.1,3,14
Furthermore, smaller NPs and metal cations can become internalised inside the bacteria,1,3 adding far-reaching antimicrobial effects, such as inhibition of enzymes crucial for DNA replication and adenosine triphosphate (ATP) production.3,14
Non-antimicrobial NPs can be useful for designing diagnostic and therapeutic strategies for bacterial infections, by adapting them for the transport-delivery of antimicrobial compounds to specific sites (nanocarriers),2,4 accumulating the compound on the bacterial surface and improving its pharmacokinetics.3,4,13,15 In this review, we discuss some examples of selective aptamers for pathogenic bacterial species which have been used with NPs for the design of novel diagnostic and/or therapeutic strategies against bacterial pathogens (Table 1).
Use of aptamers and nanoparticles used for the design of diagnostic and/or therapeutic strategies against bacterial pathogens.
| Nanostructure | Recognition element | Target bacteria | Potential utility | References |
|---|---|---|---|---|
| QDs | Aptamer | E. coli | Diagnosis | |
| Aptamer | S. typhimurium | Diagnosis | 17 | |
| Aptamer | B. subtilis | Diagnosis | ||
| Aptamer | P. aeruginosa | Diagnosis | 18 | |
| CDs | Aptamer | S. typhimurium | Diagnosis | 10,19 |
| AuNPs | Aptamer | S. enteritidis | Diagnosis | 10 |
| Aptamer | S. typhimurium | Diagnosis | 7,10,19,20 | |
| Aptamer | E. coli | Diagnosis | 7,10,19 | |
| Aptamer | S. aureus | Diagnosis | 19,36 | |
| Aptamer | C. jejuni | Diagnosis | 37 | |
| Aptamer | C. coli | Diagnosis | 37 | |
| Aptamer | L. acidophilus | Diagnosis | ||
| Au-silica NPs | Aptamer | S. typhimurium | Diagnosis | 12,21,22 |
| Aptamer | P. aeruginosa | Diagnosis | ||
| MNPs | Aptamer | S. typhimurium | Diagnosis | 23 |
| MBs and AuNPs | Aptamer | S. aureus | Diagnosis | 7,19,21,24,25 |
| Aptamer | S. typhimurium | Diagnosis | 10,26,27 | |
| MNPs and UCNPs | Aptamer | S. aureus | Diagnosis | 10,26,27 |
| Aptamer | V. parahaemolyticus | Diagnosis | 10,27 | |
| MNPs | FAM-labelled aptamer | S. typhimurium | Diagnosis | 5,10 |
| MNPs and nano gatekeepers bound to FAM-labelled oligonucleotides | Aptamer | S. aureus | Diagnosis | 28 |
| MBs, AuNCs and vancomycin | Aptamer | S. aureus | Diagnosis | 29 |
| MBs | FITC-labelled antibody and aptamer | MRSA | Diagnosis | 30 |
| SWNTs | Modified DNAzyme aptamer | S. Paratyphi A | Diagnosis | 31 |
| FNPs | Aptamer | E. coli | Diagnosis | 32 |
| FAM-labelled aptamer | V. parahaemolyticus | Diagnosis | ||
| CNPs | Cy3-labelled aptamer | S. aureus | Diagnosis | 10,33 |
| ROX-labelled aptamer | S. typhimurium | Diagnosis | ||
| AuNCs, vancomycin and AuNPs | Aptamer | S. aureus | Diagnosis | 34 |
| AuNPs and UCNPs bound to aptamer cDNA | Aptamer | E. coli | Diagnosis | 35 |
| AgNPs | fGmH-modified aptamer | S. aureus | Treatment | 40 |
| Nano gatekeepers with vancomycin | Aptamer | S. aureus | Treatment | 41 |
| TiO2NPs | Three aptamers | E. coli | Treatment | 42 |
| AuNPs coupled to A3-APOHis | Histidine-labelled aptamer | S. typhimurium | Treatment | 43 |
A3-APOHis: hexahistidine-labelled antimicrobial peptides; cDNA: complementary DNA; AgNP: silver nanoparticles; AuNCs: gold nano-clusters; AuNPs: gold nanoparticles; CDs: carbon dots; CNPs: carbon nanoparticles; CPX: ciprofloxacin; Cy3: cyanine 3 dye; FAM: carboxyfluorescein; fGmH: 2′-F-dG, 2′-OMe-dA/dC/dU; FITC: fluorescein isothiocyanate; FNPs: fluorescent nanoparticles; MBs: magnetic beads; MNPs: magnetic nanoparticles; MRSA: methicillin-resistant S. aureus; NPs: nanoparticles; QDs: quantum dots; ROX: 6-carboxy-X-rhodamine; SWNTs: single-walled carbon nanotubes; TiO2NPs: titanium dioxide nanoparticles; UCNPs: upconversion nanoparticles.
Aptamers targeting various bacterial species have been selected,1,7,8,10,12 mainly used as stationary phase for molecule capture, and are capable of identifying bacteria in environmental and clinical samples, with sensitivities equivalent to or greater than those of conventional cultures.8,12 The potential for aptamers and NPs to be used in biomedicine has been demonstrated by the synergy between them16; the affinity of an aptamer for its target is increased by a high density of aptamers on NPs, increasing the number of interactions with the target thanks to a cooperative action (multivalent effect),7,16 which in turn protects the aptamers from nuclease digestion.16
Bacteria detection based on aptamers and quantum dotsQuantum dots (QDs) are a type of NP with appreciable fluorescent properties.10,17 They have been used in pilot tests for the development of a semiquantitative detection system for Escherichia coli, Salmonella typhimurium and Bacillus subtilis by way of conjugating aptamers to QDs, and the system proved to be capable of recognising each microorganism from variations in the fluorescence of the QDs. The initial detection was evaluated with ∼2.8 × 106 bacteria/mL and the fluorescence intensity was modified in proportion to the number of bacteria present,17 showing its potential for bacterial diagnosis.18 An anti-Pseudomonas aeruginosa aptamer has also been used to develop a method for detecting this bacteria in drinking water. By labelling aptamers with fluorescein isothiocyanate (FITC) and using aptamer-conjugated QDs, these conjugates were found to have decreased affinities for the FITC-labelled aptamer,19 showing that nanotechnology can also produce poor results for promising recognition molecules.
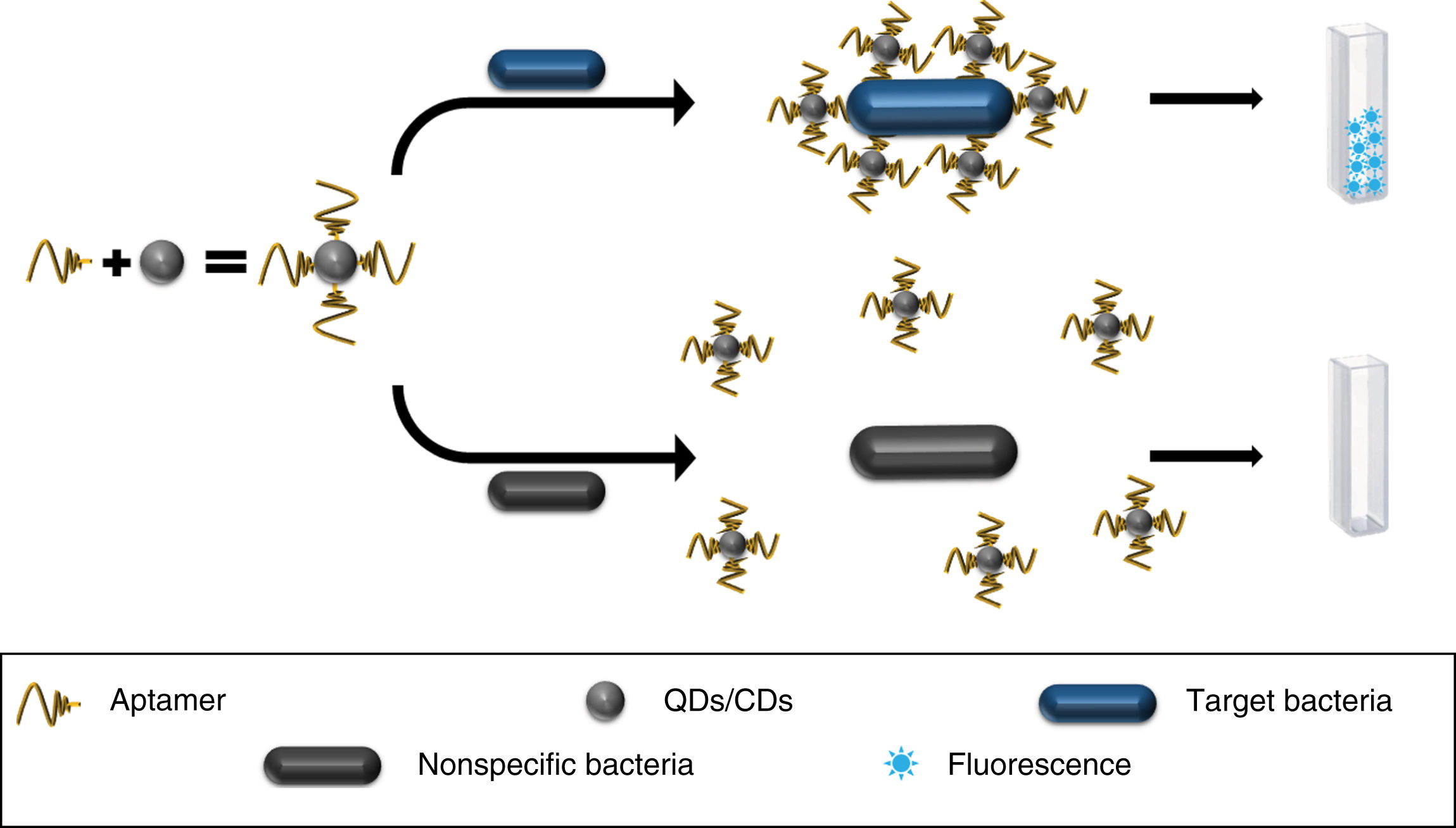
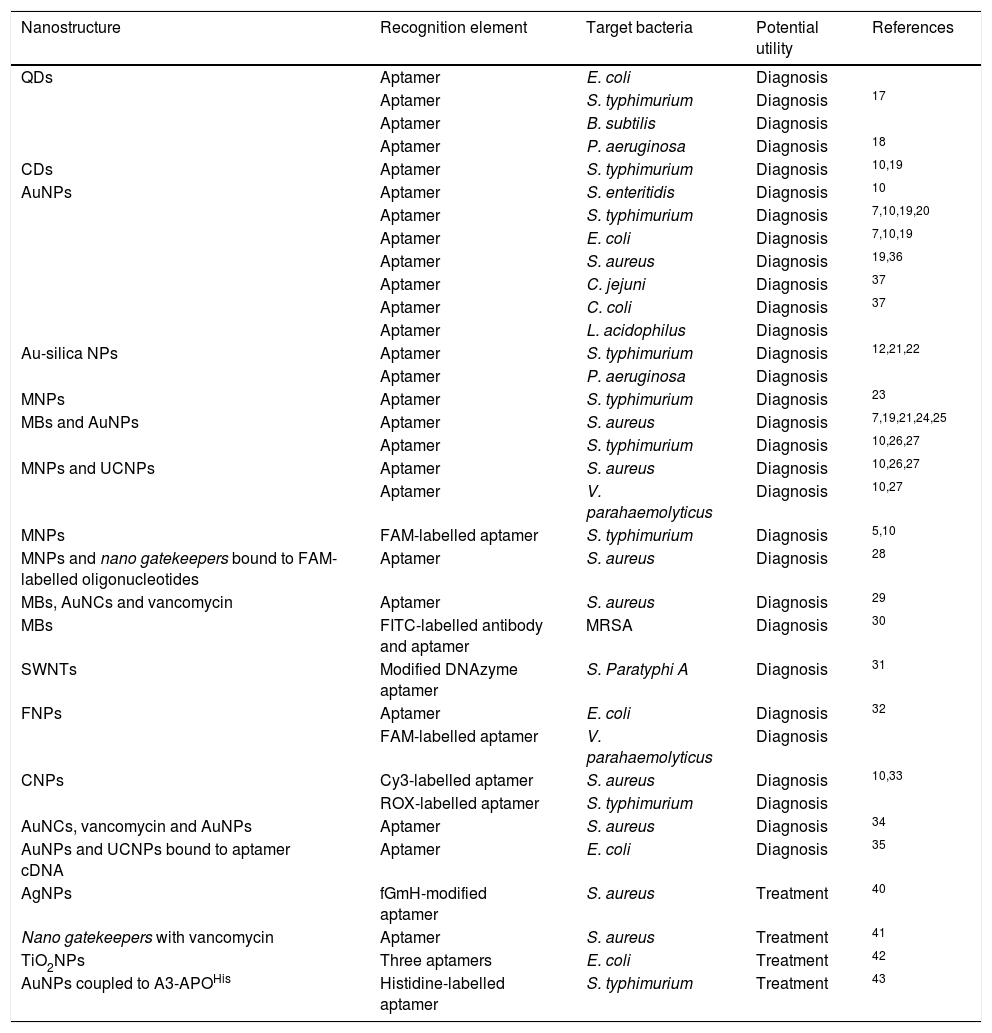
Carbon dots (CDs), with their promising luminescent, toxic and biocompatibility properties, have also been tested.10,20 They have been used in conjunction with anti-S. typhimurium aptamers to develop a fluorescence-based detection method, showing limits of detection (LOD) of 50 colony forming units (CFU)/mL in two hours of incubation with liquid bacterial cultures10,21,22 (Fig. 1), confirmed by the plate count method.22
Bacteria detection through aptamers coupled to metal nanoparticlesCurrently, gold NPs (AuNPs) are widely used for the generation of bacterial bio-sensors due to the electrochemical, optical and resonance characteristics of plasmons, among other promising properties they possess.21 They have been used in conjunction with aptamers for the detection of Salmonella enteritidis, conjugating aptamers with AuNPs, to later immobilise them on a carbon electrode. This proof of concept demonstrated that when introducing the electrode in solutions with the bacteria, the electrical resistance increased due to the formation of aptamer-bacteria complexes, allowing it to be measured by electrochemical impedance with an LOD of 600 CFU/mL.10,23 This has established the bases for using this principle to develop new methods for the detection of different microorganisms.23
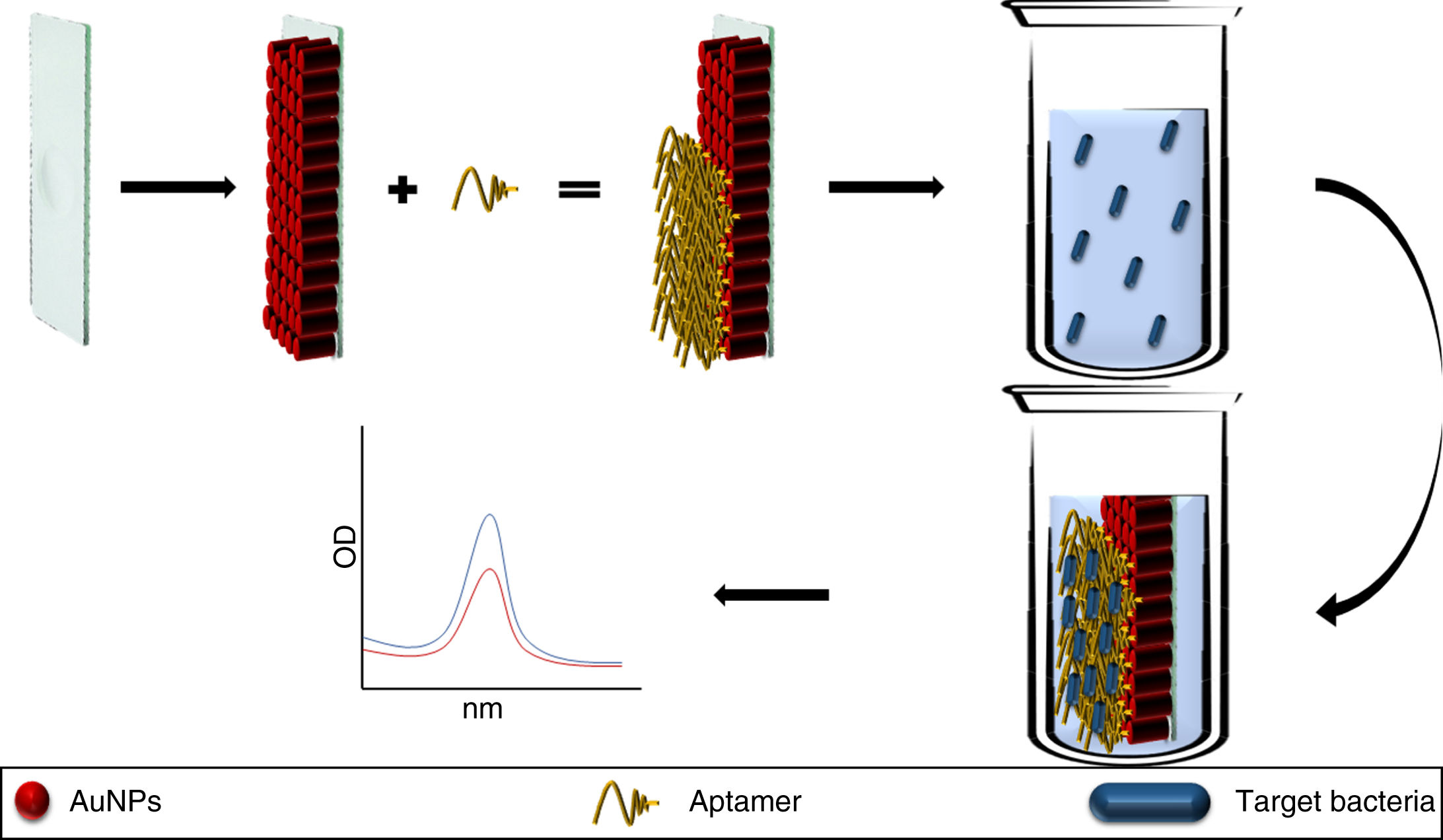
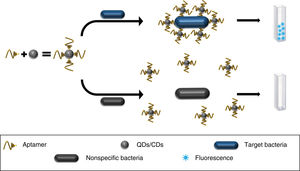
A chip has also been developed based on the immobilisation of AuNP coupled to anti-S. typhimurium aptamers, the pilot study for which showed its utility for detecting this pathogen in fluid from rinsing pork. Introducing the chip into the fluid, the aptamer changed structure on binding to the bacteria, modifying the baseline absorbance of the LSPR of the AuNPs which was detected by UV/visible spectrophotometry (Fig. 2). The main limitation of this method and this type of technology is the need to establish different matrices for different foods before it can be marketed.24
Aptamers conjugated to AuNPs and immobilised on a glass plate (chip). The aptamer binds to the target bacteria modifying the absorption peaks of the AuNPs. In solutions without the pathogen the absorption peaks remain identical to the baseline AuNP absorption peak.
AuNP: gold nanoparticles.
Another pilot study based on aptamers immobilised on silica-coated gold NPs (Au-silica NPs) enabled the development of a multiplex sensor for Lactobacillus acidophilus, S. typhimurium and P. aeruginosa, which was able to discriminate between each pathogen with an LOD of 3 CFU per assay, thanks to each aptamer recognising its target, in particular altering the LSPR of the NPs for each bacterial species.12,25,26
Aptamers and magnetic molecules for bacteria detectionMagnetic NPs (MNPs) can cause a solution to change colour in the presence of a colorimetric substrate such as 3,3′,5,5′-tetramethylbenzidine (TMB) and H2O2, in a similar way to peroxidase. This property was used to detect S. typhimurium in a proof-of-concept study, in which anti-S. typhimurium aptamers in solution with MNPs were found to inhibit MNP enzymatic activity, but on addition of 7.5 × 105 CFU/mL of the bacteria, the aptamer became bound to the pathogen, leaving the MNP unprotected and allowing their enzymatic activity.27
In addition, magnetic beads (MBs) and the MNPs can be used with the aptamers to develop methods for magnetic capture-separation of pathogens present in a sample, to concentrate them7,21,25,28–30 and subsequently detect them by various strategies, achieving LOD of 1–682 CFU/mL validated by the plate count method,29,31–33 as follows:
- (i)
From the variation in the electrical signal caused by the photonic excitation of AuNPs coupled to anti-Staphylococcus aureus (S. aureus) aptamers when bound to said pathogen.7,25,28
- (ii)
By the measurement of silver ions (Ag+) in solution, produced by silver NPs (AgNPs) coupled to anti-S. aureus aptamers, where the concentration of Ag+ is directly proportional to the density of the bacteria in a sample.21,29
- (iii)
By detection of the specific fluorescence of upconversion NPs (UCNPs) bound to specific anti-S. typhimurium, anti-S. aureus and anti-Vibrio parahaemolyticus aptamers.10,30,31
- (iv)
Similarly, from fluorescence detection of carboxyfluorescein (FAM)-modified aptamers selective for S. typhimurium.5,10,34
- (v)
Through the identification of specific enzymes produced by a particular pathogen, such as S. aureus micrococcal nucleases (MN) which, by the addition of nano gatekeepers, made up of FAM-labelled oligonucleotides specifically susceptible to MN, are immobilised in the pores of mesoporous silica NP (MSNs) to inhibit FAM fluorescence, but when MN production is stimulated, the oligonucleotides degrade, allowing the fluorescence emission.32
- (vi)
Through the specific interaction of antimicrobials with particular pathogens, such as vancomycin and S. aureus, behaviour which has been useful for the detection of this pathogen through incubation with gold nano-clusters (AuNCs) with fluorescent properties, which are inhibited by vancomycin, but in the presence of S. aureus, vancomycin interacts with the pathogen allowing fluorescence emission from the AuNCs.33
There is also evidence of the combined use of antibodies and aptamers for the fluorometric detection of certain microorganisms, such as methicillin-resistant S. aureus (MRSA) using MBs coated with S. aureus anti-protein A antibodies (SpA) to capture the pathogen. The method involves lysing the bacteria and incubating it in the presence of an FITC-modified anti-PBP2a aptamer (MRSA-specific protein) hybridised to three short DNA molecules to eliminate the fluorescence emission. When the aptamer binds to the PBP2a protein, the fluorescence emission of FITC is rehabilitated, achieving a LOD of 1.38 × 103 CFU/mL, confirmed by conventional microbiological methods.35
Bacteria detection based on aptamers coupled to nanostructuresModified deoxyribozyme (DNAzyme) aptamers immobilised on single-walled carbon nanotubes (SWNTs) have been used for the detection of Salmonella paratyphi A, where the aptamer-S. paratyphi A complex generates a conformational change of the DNAzyme-modified end, enabling it to form complexes with hemins (added to the solution) which, in the presence of luminol (also added to the system), catalyse the generation of chemiluminescence in the presence of H2O2, with a LOD of 103 CFU/mL.36
More elaborate systems for the detection of pathogens have also been designed, such as the optofluidic platform built to detect the fluorescent signal of anti-E. coli aptamers coupled to fluorescent NPs (FNPs), where the microflow of cultures through the system's microchannel made it possible to identify ∼100 E. coli cells per second by means of the fluorescent signal from the FNPs bound to them; these results then being confirmed by plate count.37
Multiplex systems have also been designed for the detection of pathogens, such as the immobilisation of anti-S. typhimurium, anti-V. parahaemolyticus and anti-S. aureus aptamers modified with different fluorochromes (FAM, cyanine dye 3 (Cy3) and 6-carboxy-X-rhodamine (ROX)) to detect each pathogen by fluorescence. The aptamers were immobilised on carbon NPs (CNPs), which enable the assembly of dyes inhibiting their fluorescence, but when the aptamers recognised their target pathogen they dissociated themselves from the CNPs, resulting in the emission and detection of fluorescence,10,38 with LOD of 50, 25 and 50 CFU/mL, respectively, validated by plate count.38
The relationship between vancomycin and S. aureus has also been used for the pilot study of a detection strategy involving variation in fluorescence resonance energy transfer (FRET), using AuNCs conjugated to vancomycin as an energy donor element and anti-S. aureus aptamers immobilised on AuNPs as an energy receptor element, which are the FRET-based dual recognition units (DRU-FRET). Both systems are attracted in the presence of the pathogen, causing the variation of FRET and enabling LOD of 10 CFU/mL.39
There is other evidence from pilot studies on strategies based on energy donor and acceptor elements, such as the use of anti-E. coli aptamers immobilised on AuNPs (energy acceptor element) and UCNPs coupled to an oligonucleotide of complementary DNA (cDNA) to the aptamer sequence (energy donor element), which when hybridised inhibit UCNP fluorescence production, but in presence of the pathogen, the aptamer binds it, dissociating its interaction with the UCNPs and causing the emission of fluorescence, with LOD of 3 CFU/mL.40
Strategies for bacteria detection based on aptamers not coupled to nanoparticlesAptamers and NPs can be used unconjugated for the detection of specific microorganisms. Some properties, such as the natural interaction between aptamers and AuNPs, have been useful in designing detection methods for E. coli, S. typhimurium and S. aureus. Proof-of-concept studies have shown that free aptamers inhibit the aggregation of AuNPs, but in the presence of the target pathogens, the aptamers bind to the bacteria, allowing the aggregation of the AuNPs (causing a change in the colour of the solution).7,10,21,41
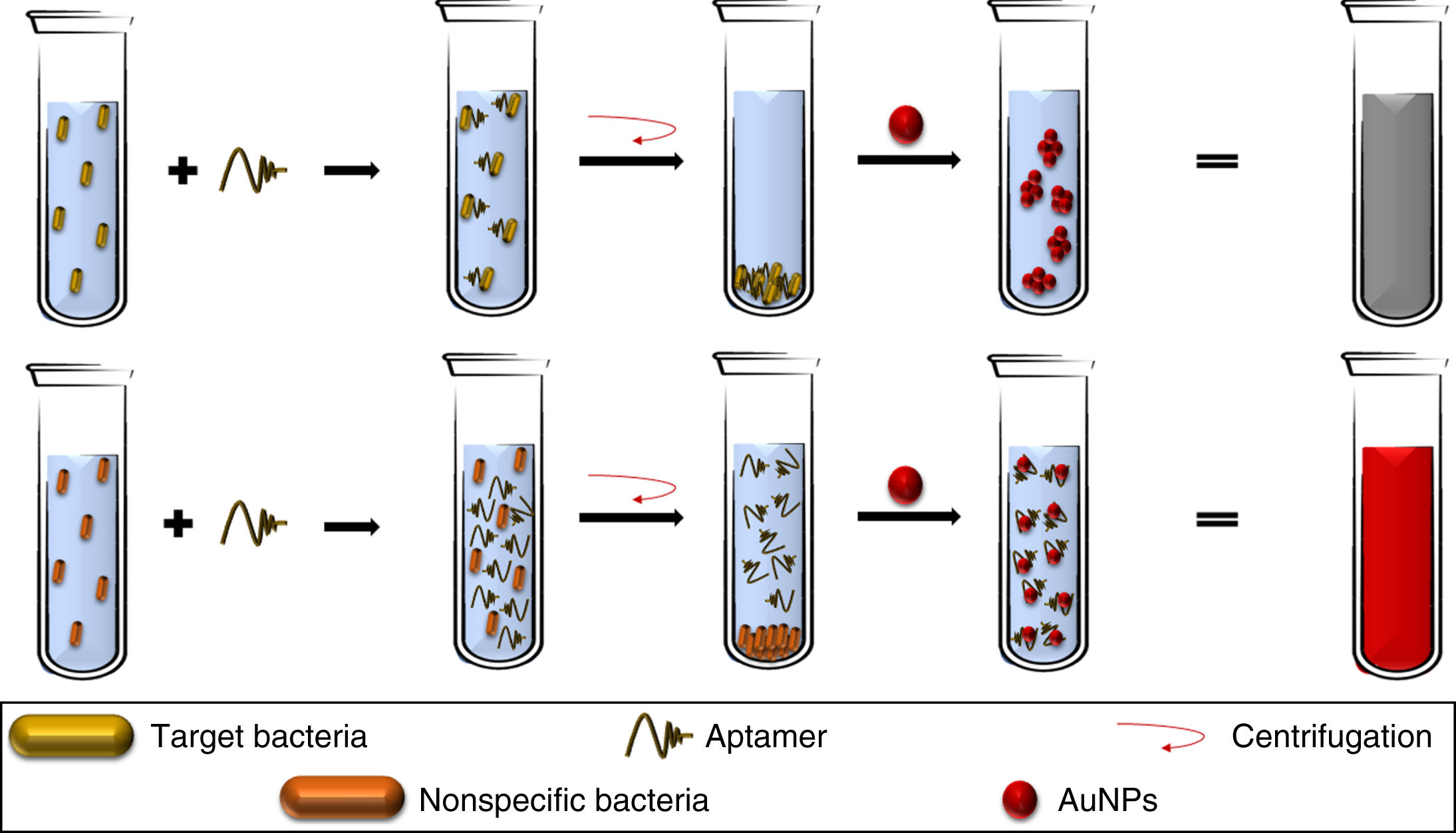
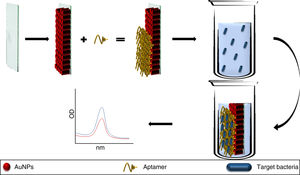
Aptamers can also be used as elements for the recognition and capture of microorganisms, as in the detection methods described for S. aureus,42Campylobacter jejuni and Campylobacter coli43 where, when added to bacterial solutions, the aptamers bind specifically to their target pathogen, being eliminated by discarding the cell button and allowing the aggregation of the AuNPs (added to the supernatant), obtaining LOD of 5.6 × 105 CFU/mL (Fig. 3); sensitivity and specificity were 80.0% and 93.3%, respectively, validated by tazobactam-supplemented cultures (standard method).43 This opens up the opportunity to use similar platforms for other pathogenic bacteria, despite the difficulties in recognising different morphological variants of the same bacterial species.43
The aptamers added to a bacterial broth specifically bind to the target bacteria. Therefore, with the cell button recovered by centrifugation, there are no free aptamers in the solution, allowing the aggregation of AuNPs. In contrast, a lack of the specific bacteria allows the presence of free aptamers, which keep the AuNPs dispersed.
AuNPs: gold nanoparticles.
Despite the promising antimicrobial characteristics of NPs and the specificity of aptamers, they have not been widely used to develop therapeutic strategies. However, their use as nanocarriers for drugs is an attractive strategy2,44,45 to increase the efficiency of the available antibiotics.45
NPs can be administered by different routes44 and at present there are antibacterial treatments such as PolyMemSilver®, Acticoat™, SilvaSorb™ and Aquacel®Ag2, to mention just a few, where AgNPs feature as one of the main elements.
There have been reports of possible toxicity of certain NPs on the host cells, although they can be modified to reduce this adverse effect.14 One example is conjugation of the NPs with aptamers,45 opening up the possibility of delivering NPs and/or other drugs to the right place at the right concentrations and for the right amount of time,2 which increases their antimicrobial power in the range of 3- to 250-fold,2,14 and this is why they have been useful for the development of novel therapeutic strategies against different types of cancer.15
In vitro studies on strategies with therapeutic potentialIn terms of treatment against bacterial infections based on aptamers and NPs, the evidence remains scarce but is encouraging. One possibility is the modification of an anti-SpA aptamer with fGmH (2′-F-dG, 2′-OMe-dA/dC/dU), giving it resistance to alkaline hydrolysis and to nucleases present in serum. Additionally, the aptamers conjugate with AgNPs, releasing their specific antimicrobial action against S. aureus in an SpA-dependent manner.46
Nano gatekeepers have been found to be effective as nanocarriers for antibiotics; for example, immobilisation of vancomycin in the pores of MSNs and its subsequent conjugation with an anti-S. aureus aptamer, delivering the drug to the exact site and thereby reducing its minimum inhibitory concentration and its toxicity against other related species.47
Moreover, the antimicrobial effect of NPs can be improved with the use of different aptamers targeted against the same pathogen, as described for E. coli, where the immobilisation of three aptamers on titanium dioxide NPs (TiO2NPs) deactivated 99.9% of bacteria in 30 min, in contrast to TiO2NPs bound to one aptamer and TiO2NPs alone, which deactivated the bacteria in 60 min.48
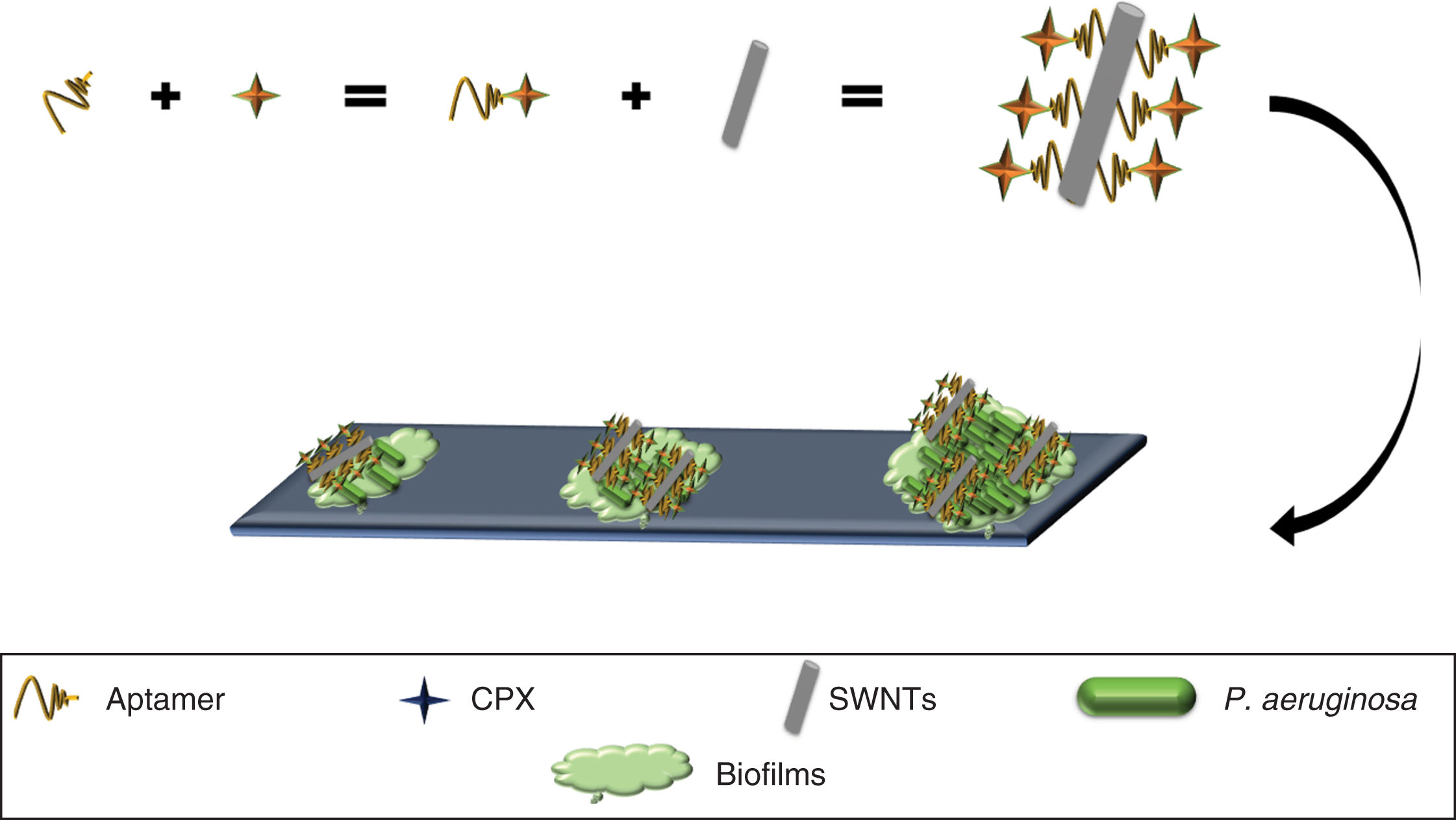
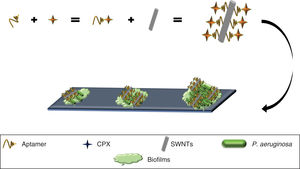
SWNTs have been shown to possess activity against biofilms49 (primary strategy used by bacteria to survive in different environments)50 and have been used to develop an anti-biofilm nanocomposite of P. aeruginosa, by using a selective aptamer against this bacteria conjugated to ciprofloxacin (Apt-CPX) and SWNTs (Apt-SWNTs). Additionally, a molecule was generated made up of the three elements (Apt-CPX-SWNTs) which, in vitro, was able to reduce biofilm formation by 90% and degrade ∼75% of established biofilms (Fig. 4), indicating that these tools could be used for the effective treatment of biofilms.49
In vivo trials of bacterial eradication therapiesThere have been promising results with histidine-labelled anti-S. typhimurium aptamers coupled to AuNPs (AuNP-AptHis), a complex which is in turn conjugated to hexahistidine-labelled antimicrobial peptides (A3-APOHis), in eradicating intracellular infections by S. typhimurium; they have been shown to work in vitro by releasing A3-APOHis inside HeLa cells infected with the bacteria. In in vivo assays on mice infected with doses that caused the death of the animal in 4–5 days, the molecule enabled its survival by eradicating the pathogen, verified by a decrease of ∼93−98% in the number of viable bacterial cells in cultures from the infected organs of each mouse,51 suggesting that the combined use of aptamers, NPs and even other antimicrobial compounds may be useful for the development of effective therapies against infections caused by bacteria.
ConclusionsIn recent years, nanometric biomedicine has been positioning itself as a promising tool for the diagnosis, prevention and treatment of a number of different diseases, where aptamers and NPs have demonstrated their applicability for diagnosis and treatment. In bacterial infections, proofs of concept of the combined use of both elements have enabled the rapid and specific detection of individual bacterial cells. The future implementation of these methods could therefore enable accurate diagnoses, improving the prognosis of infected patients by allowing them to receive early, specific therapy for the eradication of the infectious agent. Furthermore, in vivo studies of complex nanostructures consisting of NPs, aptamers and even other antimicrobial compounds have shown that they could be a powerful tool in the face of the “antibiotic resistance crisis”, allowing eradication of infections with low doses of antimicrobials by delivering them multivalently to the right place and for the precise amount of time, being an ideal therapy which would not affect the organ or tissue microbiota or the host cells. These technological advances suggest that there may soon be rapid and specific tools based on NPs and aptamers for the diagnosis and treatment of infectious diseases.
FundingJuan Carlos Gutiérrez Santana is beneficiary of the 2018-000068-02NACF-28106 grant from the Consejo Nacional de Ciencia y Tecnología (CONACyT) [National Council of Science and Technology], Mexico, and would like to thank this body for its support. This work was financed by federal funds from the Instituto Nacional de Pediatría [National Institute of Paediatrics] in Mexico, authorisation 068/2019.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Gutiérrez-Santana JC, Toscano-Garibay JD, López-López M, Coria-Jiménez VR. Aptámeros acoplados a nanopartículas para el diagnóstico y tratamiento de las infecciones microbianas. Enferm Infecc Microbiol Clin. 2020;38:331–337.