The carbapenem inactivation method (CIM) is a cost-effective assay for detecting carbapenemases. However, its interpretation is unclear for Pseudomonas spp. We evaluate its accuracy when meropenem is changed to imipenem.
MethodsWe analyzed 266 P. aeruginosa isolates. The CIM method consists of: resuspend bacterial colonies (a full 10μL loop) in 400μL water, in which a 10μg disk of meropenem/imipenem is immersed. After 2h of incubation (35°C), remove the disk, place it onto a Mueller-Hinton agar plate previously inoculated with Escherichia coli (ATCC 25922), and incubate at 35 ̊C between 18-24 h. Interpretation criteria (mm of inhibition zone): ≤19mm, positive; ≥25mm negative; 20–24mm, undetermined.
ResultsImipenem improves the sensitivity and specificity of CIM when compared to meropenem (99.4% and 98.9%, vs. 91.9% and 94.7%, respectively).
ConclusionsThe accuracy of CIM for carbapenemase detection in P. aeruginosa is increased with the use of imipenem.
El método de inactivación del carbapenem (CIM, por sus siglas en inglés) se utiliza para detectar carbapenemasas, pero su interpretación no está clara para Pseudomonas spp. Se evaluó la precisión del método utilizando imipenem en lugar de meropenem.
MétodosSe estudiaron 266 aislados de P. aeruginosa. El CIM consiste en: suspender colonias bacterianas (un asa de 10μl) en 400μl de agua en los que se sumerge un disco de 10μg de meropenem/imipenem. Tras 2h de incubación (35°C) se saca el disco y es transferido a agar Mueller-Hinton, previamente inoculado con Escherichia coli (ATCC 25922) e incubado a 35°C entre 18-24h. Criterios de interpretación (mm halo de inhibición): ≤19mm positivo; ≥25mm negativo y 20-24mm indeterminado.
ResultadosImipenem mejora la sensibilidad y especificidad del CIM frente a meropenem (99,4 y 98,9% vs. 91,9 y 94,7%, respectivamente).
ConclusionesLa precisión del CIM para la detección de carbapenemasas en P. aeruginosa mejora al utilizar imipenem.
The global spread and increasing prevalence of multidrug-resistant (MDR) bacteria is a severe public health problem.1Pseudomonas aeruginosa is an important nosocomial pathogen, which can become resistant to antibiotics due to low permeability, expression of various efflux pumps, and the production of β-lactamases.2 Carbapenemase production in P. aeruginosa is the most threatening mechanism of resistance since carbapenemases are able to hydrolyze almost all β-lactam antibiotics and are easily transferable to other bacteria through plasmids.2 The most common carbapenemases reported in P. aeruginosa are metallo-β-lactamases (MBLs) such as VIM, IMP, SPM, and Ambler class A carbapenemases like KPC.3 Screening for carbapenemases is essential for establishing appropriate infection control measures.2
Currently, several phenotypic methods are available for detecting carbapenemases such as rapid colorimetric tests (Carba NP test and Blue-Carba assay), the inhibitor-based methods (ethylenediaminetetraacetic acid (EDTA), sodium mercaptoacetate (SMA) and dipicolinic acid (DPA), immunochromatographic assays, the carbapenem inactivation method (CIM), the modified carbapenem inactivation method (mCIM) and the mCIM which uses Tris–HCl, called CIMtris.4 It should be noted that detecting carbapenemases is more difficult in P. aeruginosa compared to Enterobacteriaceae.1 The CIM is a novel, simple and low-cost phenotypic method, modified and recommended by the Clinical and Laboratory Standards Institute (CLSI) for the detection of carbapenemases in Enterobacteriaceae and non-fermentative bacteria.5 Several published studies have reported high sensitivities and specificities around 99% using the CIM in Enterobacteriaceae.6,7 However, there is a limited number of studies which have evaluated the CIM performance in non-fermentative bacteria.8–12 Seeking an efficient way to detect carbapenemases in non-fermentative microorganisms (P. aeruginosa) with the CIM, the aim of this study was to determine whether the accuracy in detecting carbapenemases could be higher when changing the substrate from meropenem disks (as recommended by the CLSI) to imipenem disks and implementing the method with a measurable interpretive criteria based on the size (mm) of the inhibition zone.
MethodsWe used a random sample including 266 carbapenem resistant5P. aeruginosa strains collected between 2009 and 2017 from Colombian hospitals belonging to a bacterial resistance surveillance network. All isolates had been previously characterized using a real-time multiplex PCR to identify carbapenemase genes (blaVIM, blaIMP, blaKPC, blaNDM) following previously reported conditions.13 Among the 266 strains, 171 strains produced carbapenemases: KPC (n=67), VIM (n=91) and KPC-VIM co-producers (n=13) and 95 strains were non-carbapenemase producers. In order to determine if there was a higher accuracy by changing the carbapenem substrate, we used and compared meropenem (10μg) and imipenem (10μg) (Oxoid Ltd, Hampshire, United Kingdom) disks.
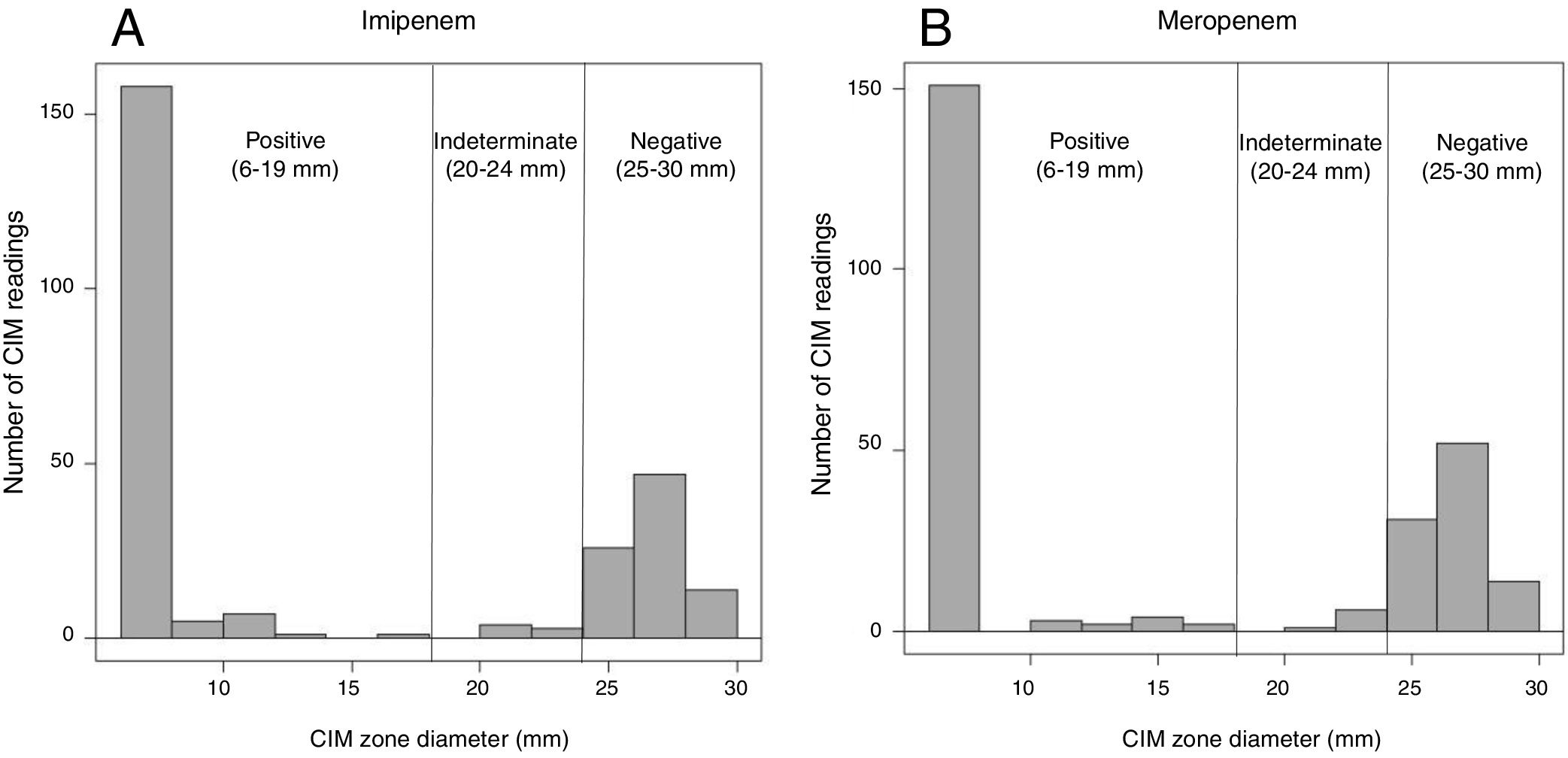
The CIM was implemented as described by Van Der Zwaluw et al.9 Briefly, the CIM was performed as follows: a full 10μL inoculation loop of the culture taken from a Mueller-Hinton or blood agar plate was suspended in 400μL of water in an Eppendorf tube. Then, a disk containing 10μg of meropenem/imipenem was immersed in the suspension and incubated at 35°C. After 2h of incubation, the disk was removed from the suspension using an inoculation loop and placed onto a Muller-Hinton agar plate (BioMeriéux, France) previously inoculated with a susceptible Escherichia coli (ATCC 25922) and subsequently incubated overnight at 35°C. During the procedure, the carbapenem disk was wholly immersed in the bacterial suspension (not on the surface of the suspension), and additional vortexing was performed. After the incubation, the inhibition zone was measured and we established measurable interpretive criteria based on the mm of inhibition. The measurable criteria for interpretation, was defined as follows: <=19mm was considered positive, >= 25mm negative, and 20–24mm was considered undetermined (Fig. 1). This measurable criterion was established taking into account the average of inhibition zones obtained for carbapenemase-producers and for non-carbapenemase-producers (according to PCR results and the CIM).
Distribution of carbapenem inactivation method (CIM) zone diameter measurements. Displayed are the number of CIM readings with specific zone diameter measurements during the study. A total of 266 isolates were tested, 171 were carbapenemases-producers and 95 were non-carbapenemases producers. (A) Zone diameter measures representing the data obtained using imipenem disk as substrate in the CIM; (B) zone diameter measures representing the data obtained using meropenem disk as substrate in the CIM.
The strains were tested in duplicate. Control strains were ATCC 27853 P. aeruginosa as a negative control, and a carbapenemase-producing P. aeruginosa (IMP-1) clinical isolate characterized by molecular methods as a positive control. Also we performed the manual CARBA NP method with the purpose of confirming the non-carbapenemase producers. Sensitivity, specificity, positive predictive value and negative predictive value, were calculated in order to determinate the accuracy of the CIM versus the results obtained by PCR that was used as the gold-standard result. All calculations were performed using the statistical program R (Version 3.3.2).
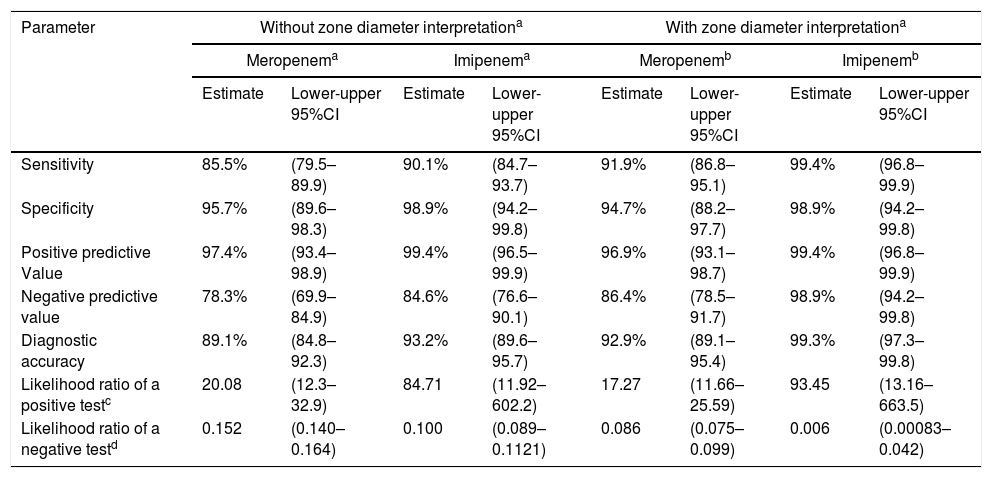
ResultsA summary of data is presented in Table 1. A total of 171 isolates were carbapenemase-producers, and 95 were non-carbapenemase producing isolates. By using meropenem disk results interpretation, based on the absence or presence of inhibition zones, 25 carbapenemase-producers were not detected out of 171 analyzed (9 isolates with KPC, 12 with VIM, and 4 with KPC and VIM). By the contrary, four non-carbapenemase producing isolates were detected as positive with the test. The sensitivity was 85.5% (95%CI 79.5–89.9) and the specificity 95.7% (95%CI 89.6–98.3). Simultaneously, when using imipenem disks and the same interpretive criteria, 17 carbapenemase-producers were not detected (12 isolates with VIM, 1 with KPC and 4 with KPC and VIM) and one non-carbapenemase producer was positive with the test. The sensitivity and specificity were 90.1% (95%CI 84.7–93.7) and 98.9% (95%CI 98.9–99.8%) respectively.
Values obtained with the carbapenem inhibition method (CIM) in P. aeruginosa isolates (n=266).
| Parameter | Without zone diameter interpretationa | With zone diameter interpretationa | ||||||
|---|---|---|---|---|---|---|---|---|
| Meropenema | Imipenema | Meropenemb | Imipenemb | |||||
| Estimate | Lower-upper 95%CI | Estimate | Lower-upper 95%CI | Estimate | Lower-upper 95%CI | Estimate | Lower-upper 95%CI | |
| Sensitivity | 85.5% | (79.5–89.9) | 90.1% | (84.7–93.7) | 91.9% | (86.8–95.1) | 99.4% | (96.8–99.9) |
| Specificity | 95.7% | (89.6–98.3) | 98.9% | (94.2–99.8) | 94.7% | (88.2–97.7) | 98.9% | (94.2–99.8) |
| Positive predictive Value | 97.4% | (93.4–98.9) | 99.4% | (96.5–99.9) | 96.9% | (93.1–98.7) | 99.4% | (96.8–99.9) |
| Negative predictive value | 78.3% | (69.9–84.9) | 84.6% | (76.6–90.1) | 86.4% | (78.5–91.7) | 98.9% | (94.2–99.8) |
| Diagnostic accuracy | 89.1% | (84.8–92.3) | 93.2% | (89.6–95.7) | 92.9% | (89.1–95.4) | 99.3% | (97.3–99.8) |
| Likelihood ratio of a positive testc | 20.08 | (12.3–32.9) | 84.71 | (11.92–602.2) | 17.27 | (11.66–25.59) | 93.45 | (13.16–663.5) |
| Likelihood ratio of a negative testd | 0.152 | (0.140–0.164) | 0.100 | (0.089–0.1121) | 0.086 | (0.075–0.099) | 0.006 | (0.00083–0.042) |
Accuracy values according with the following interpretation. Measure of the inhibition zone: 6mm (positive); >6mm (negative). Mean of values not applying zone diameter interpretation)
Accuracy values according with the following interpretation. Measure of the inhibition zone: 6–19mm (positive); >25mm (negative); 20–24 (indeterminate). Mean of values applying zone diameter interpretationa.
When using the measurable criteria, for meropenem, the false negative results were lower (14 carbapenemases-producers were negative), the sensitivity increased to 91.9% (95%CI 86.8–95.1) and the specificity was 94.7% (95%CI 88.2–97.7). On the other hand, for imipenem, the false negative results were significantly lower (only one carbapenemase-producer was negative), with sensitivity increasing to 99.4% (95%CI 96.8–99.9), and the specificity to 98.9% (95%CI 94.2–99.8). Therefore, the study showed that the likelihood ratios for a positive test (17.27–93.45) and the likelihood ratios for a negative test (0.006–0.152) when using the CIM were excellent, and particularly when imipenem was used as a substrate.
DiscussionThe acquisition of carbapenemase genes by P. aeruginosa is an important cause of MDR, and therefore, the rapid and correct detection of carbapenemases is crucial.1 Molecular methods are the gold standard in the identification of carbapenemase-producing strains,1 but phenotypic methods have been developed due to the high cost of molecular methods and their inability to detect new carbapenemase genes.2 However, some phenotypic assays are still not accepted for non-fermentative Gram-negative bacilli.1 The mCIM for Pseudomonas spp. has been included in the CLSI guideline in 2018.7 This mCIM consists of a modification in which TBS broth is used instead of water and the incubation time is prolonged from 2h to 4h.5 Van der Zwaluw et al.,9 reported that the concordance with the molecular method was 97% in Pseudomonas spp. Simner et al. described a sensitivity and specificity of 79% and 100%, respectively, for the CIM method, and a sensitivity of 100% and specificity of 98% for the mCIM method.11
We demonstrate that the use of imipenem as the substrate in the CIM method, instead of meropenem for the detection of carbapenemases increases both the sensitivity and the specificity (sensitivity of 90.1% and specificity of 98.9% for imipenem versus sensitivity of 85.5% and specificity of 95.7% for meropenem). Additionally, the high sensitivity and specificity as well as the low-cost and simplicity of use, make this test a very promising tool to be implemented routinely. Furthermore, the measure of the inhibition zones by using the disks makes interpretation easier and also enhances both the sensitivity and specificity (sensitivity of 99.4% and specificity of 98.9% for imipenem versus sensitivity of 91.9% and specificity of 94.7% for meropenem).
Besides, interpreting the results of the CIM is more precise than for other assays. The main limitation of the CIM is the time required to obtain results (18–24h). Uechi et al. reported for their CIMtris modification an overall sensitivity of 97.6% and specificity of 92.6% in comparison to mCIM (sensitivity of 45.1% and specificity of 100%) in Pseudomonas spp.4 Another limitation of the CIM test is that it does not differentiate the type of carbapenemase produced, which is essential since the new therapeutic options for the treatment of infections due to carbapenemase-producing organisms such as ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam are active against some but not all types of carbapenemases.14
The main limitation of this study is the small number of carbapenemase-types tested, which do not reflect the global epidemiology of carbapenemases. However, our data reflect the epidemiological situation in our region, since our isolates belong to collections obtained in the routine microbiology laboratory. We believe that the CIM for P. aeruginosa is an accurate, useful, quick, inexpensive and simple tool that should be incorporated in countries with limited resources. On the other hand, the CIM is the most practical assay to select since all the necessary supplies are readily available in comparison to the use of the mCIM5 that requires more specific reagents which are not available in all clinical laboratories in Latin America. Our findings only suggest an improved option of a phenotypic assay that should be used for detecting carbapenemase-producing P. aeruginosa isolates especially in clinical microbiology laboratories with limited resources.
FundingThis study was supported by COLCIENCIAS in the framework of the project number 222971250821 and was partially funded byMerck Sharp & Dohme, and Pfizer, which help fund the Bacterial Resistance Study Group.
Conflict of interestsMVV and CJP have received consulting fees and/or research grants from Merck Sharp & Dohme, Pfizer, WEST and GPC pharma. CHG is currently an employee of MSD Colombia. All other authors declare no competing interests.
We thank the institutions that belong to the Colombian Nosocomial Resistance network led by the Bacterial Resistance and Hospital Epidemiology Unit, in CIDEIM, Cali, Colombia.