A description is presented on the molecular epidemiology of carbapenemase-producing enterobacteriaceae infection in a tertiary hospital.
Material and methodsA study was made on all the carbapenemase-producing enterobacteriaceae isolations obtained between February 2015 and March 2016 in the Hospital Universitario 12 de Octubre (Madrid). Phenotypic and molecular methods were used.
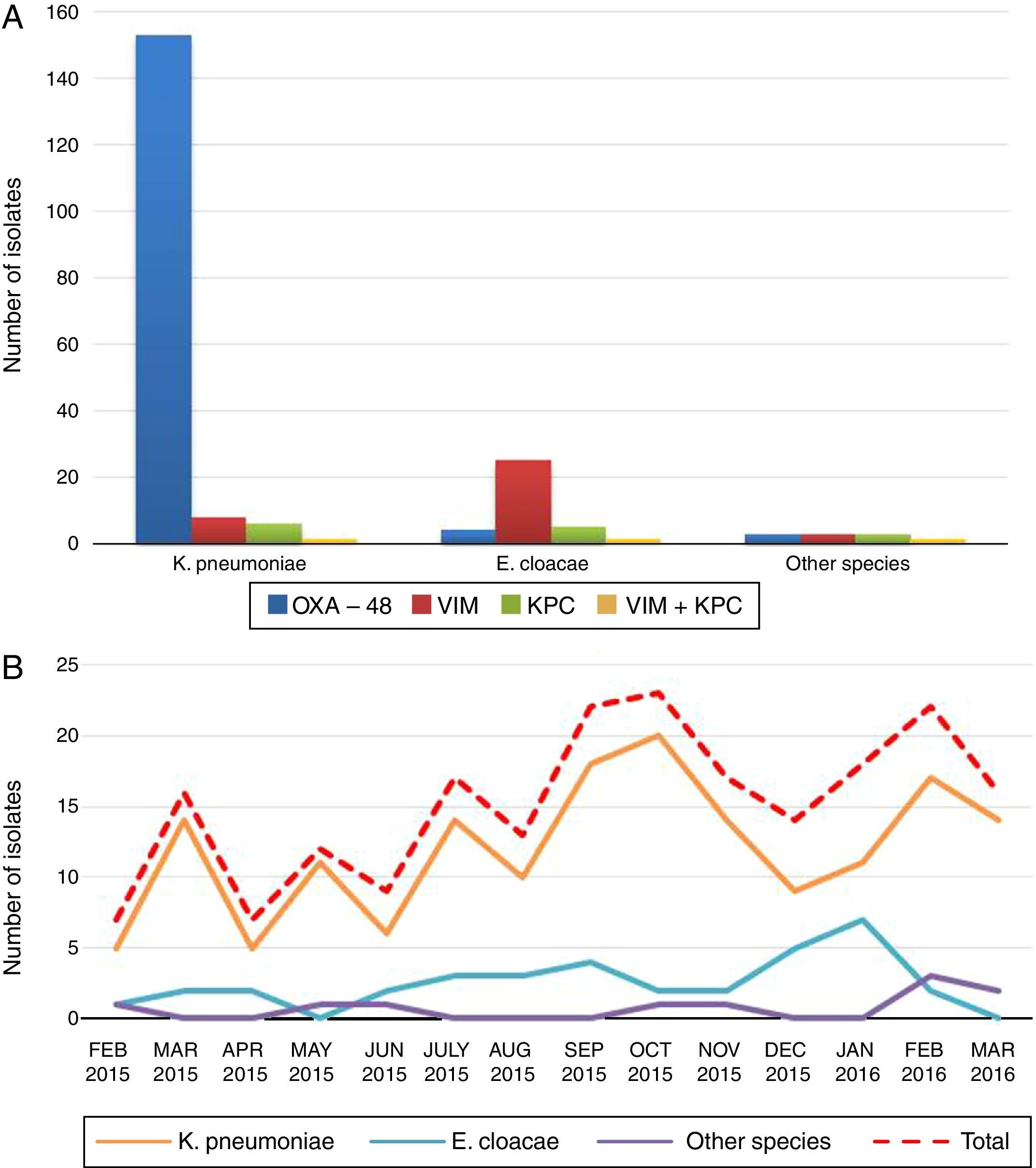
ResultsA total of 7 bacterial species were identified, with the majority being Klebsiella pneumoniae (K. pneumoniae) (78.9%) and Enterobacter cloacae (E. cloacae) (16.4%). The resistance of K. pneumoniae and E. cloacae for carbapenems was 88.7 and 88.6% for ertapenem, 21.4 and 54.3% for imipenem, and 20.8 and 34.3% for meropenem, respectively. The most frequent carbapenemase type was OXA-48 (91.1%) and VIM (71.4%) in E. cloacae. A total of 9 K. pneumoniae clonal types were identified, including a majority pertaining to the sequence type ST11. In E. cloacae, 16 clonal types were identified.
ConclusionsThe current increase in carbapenemase-producing enterobacteriaceae is mainly due to the spread of OXA-48-producing K. pneumoniae.
Se describe la epidemiología molecular de las enterobacterias productoras de carbapenemasas en un hospital terciario.
Material y métodosSe incluyeron todos los aislamientos de enterobacterias productoras de carbapenemasas obtenidos entre febrero de 2015 y marzo de 2016 en el Hospital Universitario 12 de Octubre (Madrid). Se utilizaron métodos fenotípicos y moleculares.
ResultadosSe identificaron 7 especies bacterianas, predominando Klebsiella pneumoniae (K. pneumoniae) (78,9%) y Enterobacter cloacae (E. cloacae) (16,4%). La resistencia en K. pneumoniae y E. cloacae para carbapenemes fue del 88,7 y 88,6% para ertapenem, 21,4 y 54,3% para imipenem, y 20,8 y 34,3% para meropenem. El tipo de carbapenemasa más frecuente en K pneumoniae fue OXA-48 (91,1%) y en E. cloacae VIM (71,4%). Se identificaron 9 tipos clonales de K. pneumoniae, incluyendo uno mayoritario perteneciente al tipo de secuencia ST11, y 16 de E. cloacae.
ConclusionesEl incremento actual de enterobacterias productoras de carbapenemasas se debe en gran medida a la diseminación de K. pneumoniae productora de OXA-48.
Multi-resistant bacteria infections are one of the main public health problems and in our centre, the most important infections are caused by carbapenemase-producing Enterobacteriaceae (CPE).1 In Europe, and specifically in Spain, OXA-48-like carbapenemase is the most frequent strain and is especially relevant in Klebsiella pneumoniae (K. pneumoniae).2,3 The increase in the number of CPE infections/colonisations has been caused, to a great extent, by the dissemination of epidemic clones, as in the case of K. pneumoniae.4 An earlier study conducted in our hospital (2009–2014) showed the emergence of carbapenemase producing K. pneumoniae, mainly due to the dissemination of an ST11 clone.5 Following on from this, the need arose to determine the molecular epidemiology of CPE colonisations/infections in our hospital and to investigate whether some high-risk clones persist.
Material and methodsThe study included all CPE isolates from February 2015 to March 2016 at the University Hospital 12 de Octubre (Madrid). Only 1 isolate per patient was included. Identification was carried out using the MALDI-TOF MS system (Microflex, Bruker Daltonics, Bremen, Germany). Antibiotic sensitivity testing was performed using the microdilution method (Neg Combo Panel Type 53 and Neg Urine Combo Panel Type 59, Microscan Walkaway, Soria Melguizo, Madrid, Spain) and the interpretation criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST version 5.0, 2015) were applied to determine sensitivity to antibiotics. The modified Hodge test was performed on isolates with suspected carbapenemase and the exact MIC for carbapenems (ertapenem, imipenem and meropenem) and colistin was determined using the E-test (Biomérieux, Durham, NC). Isolates that were positive for carbapenemase using the Hodge test underwent real-time PCR testing using specific primers directed against the blaOXA-48, blaVIM and blaKPC6–8 genes, which are the most frequent carbapenemase types in Spain. Sequencing was performed using the BigDye 3.1 system (3130 Genetic Analyzer, Applied Biosystems, Austin, TX) in representative samples of each type of carbapenemase to confirm the results obtained by PCR.
All strains of Enterobacter cloacae (E. cloacae) and a representative selection of strains of K. pneumoniae were studied to determine the epidemiological link between isolates from all hospital departments over the study period. Pulsed field gel electrophoresis (PFGE) was performed after digestion with the XbaI enzyme using the CHEF DRIII system (Bio-Rad Laboratories, Hercules, CA). The different band patterns were analysed using the Bionumerics software package v.3.0 (Applied Maths NV, Sint-Martens-Latem, Belgium).
ResultsDuring the 14 months of the study, a total of 213 CPE isolates were identified, 139 (65.3%) from clinical samples (urine [No.=81], exudate/wound pus [No.=24], blood [No.=13], respiratory [No.=11], organic liquid [No.=6], catheter [No.=3] and biopsy [No.=1]) and 74 (34.7%) from surveillance culture samples from multi-resistant bacteria carriers (perianal exudates). The isolates belonged to seven bacterial species: K. pneumoniae (78.9%), E. cloacae (16.4%), Citrobacter freundii–C. freundii–(1.9%), Klebsiella oxytoca–K. oxytoca–(0.9%), Escherichia coli–E. coli–(0.9%), Serratia marcescens–S. marcescens–(0.5%) and Proteus mirabilis–P. mirabilis–(0.5%) (Fig. 1, panel A). The percentage of CPE with respect to the total number of isolates, by species, was: K. pneumoniae 4.9% (168/3404), E. cloacae 4.2% (35/841), C. freundii 3.1% (4/127), K. oxytoca 0.4% (2/480), S. marcescens 0.2% (1/472), P. mirabilis 0.1% (1/864) and E. coli 0.02% (2/11298). Nearly all (94.4%) (201/213) of patients infected/colonised by these bacteria were adults, admitted to the following hospital departments: ICU (29.9%), A&E (16.5%), Internal Medicine (12.4%), Surgery (11.3%), Nephrology/Urology (9.8%) and other departments (11.3%); 8.8% were referred from Primary Care. In the case of paediatric patients (5.4%), the largest number of CPEs (9/12) were isolated from patients in the paediatric ICU. The cases occurred at an increasing rate over the entire study period (Fig. 1, panel B).
With regard to the type of carbapenemase detected, 91.1% (153/168) of the K pneumoniae isolates were OXA-48-like producers, 4.8% were VIM producers (8/168), 3.6% were KPC producers (6/168) and 0.6% (1/168) were VIM and KPC co-producers. Regarding E. cloacae, 71.4% were VIM producers (25/35), 14.3% were KPC producers (5/35), 11.4% were OXA-48 producers (4/35) and 2.9% (1/35) were VIM and KPC co-producers (Fig. 1, panel A).
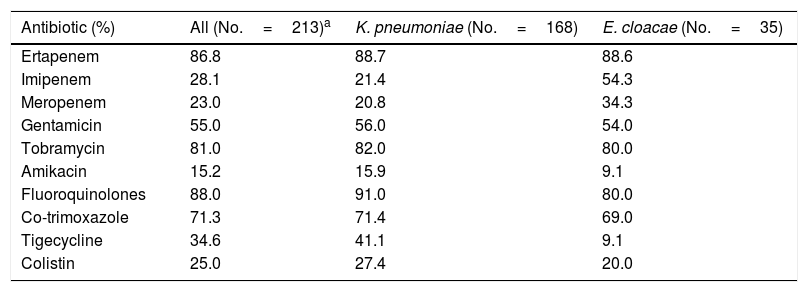
Regarding antimicrobial sensitivity testing, meropenem was the most active carbapenem, with an overall resistance of 23% among CPEs (20.8% and 34.3% in K. pneumoniae and E. cloacae, respectively) (Table 1). Analysing carbapenemase by type showed that the KPC-producing strains presented the highest percentage of carbapenem resistance (ertapenem, imipenem and meropenem: 100%, 50% and 50% in K. pneumoniae and 100%, 80% and 80% in E. cloacae, respectively). The percentages of resistance to the remaining antibiotics are shown in Table 1.
Pattern of CPE antibiotic resistance.
| Antibiotic (%) | All (No.=213)a | K. pneumoniae (No.=168) | E. cloacae (No.=35) |
|---|---|---|---|
| Ertapenem | 86.8 | 88.7 | 88.6 |
| Imipenem | 28.1 | 21.4 | 54.3 |
| Meropenem | 23.0 | 20.8 | 34.3 |
| Gentamicin | 55.0 | 56.0 | 54.0 |
| Tobramycin | 81.0 | 82.0 | 80.0 |
| Amikacin | 15.2 | 15.9 | 9.1 |
| Fluoroquinolones | 88.0 | 91.0 | 80.0 |
| Co-trimoxazole | 71.3 | 71.4 | 69.0 |
| Tigecycline | 34.6 | 41.1 | 9.1 |
| Colistin | 25.0 | 27.4 | 20.0 |
Seventy four isolates were selected for the molecular typing study, 39K. pneumoniae (36 OXA-48-like, 2 VIM and 1 KPC) and 35 E. cloacae (25 VIM, 5 KPC, 4 OXA-48 and 1 VIM-KPC). In the case of K. pneumoniae, the analysis revealed the presence of 9 clonal types, 5 patterns were found in 89.7% (35/39) of the isolates with a predominant clone (clone A) and 4 showed a unique pattern. Clone A included 18 OXA-48-producing and 2 VIM-producing isolates and was found in patients admitted to 4 departments (ICU, Surgery, Internal Medicine and A&E) and also in primary care patients. The comparison of PFGE patterns with those included in our laboratory database allowed us to identify clone A as the predominant clone in the 2009–2014 period in our hospital as well. This clone had previously been identified as ST11 (JNHB00000000). The PFGE study in E. cloacae revealed the presence of 16 clonal types; 24 isolates corresponded to 5 clonal types and 11 showed a unique pattern.
DiscussionResistance to beta-lactam antibiotics in Enterobacteriaceae has increased dramatically in recent years, mainly due to the increase in carbapenemase-producing strains.9 Our results show that up to 7 different species of Enterobacteriaceae are producers of these enzymes, most notably OXA-48 K. peumoniae. A multicentre study conducted in 2013 reported up to 9 species of CPE, the most common being OXA-48-producing K. pneumoniae.2
The molecular epidemiology study shows different patterns of CPE dissemination in our hospital. On the one hand, the emergence and spread of K. pneumoniae occurs as a result of a few, predominantly OXA-48-producing clones. The appearance of high epidemic risk clones, such as carbapenemase-producing K. pneumoniae ST11, aggravates the problem, since this clone is widely distributed in Spain and in other European countries and can contain different types of carbapenemase in addition to multiple resistance to other antimicrobial groups.4 In our hospital, it was first identified in 2011 in a VIM-1-producing strain. From then on, it extended to all clinical departments but as of 2012, most of the isolates were OXA-48 producers.5 Our results confirm the persistence of this clone and its wide distribution throughout the hospital. Despite considerable efforts to control the spread of this type of infection, it is now endemic to our institution. In contrast, E. cloacae showed a very different pattern, since 16 clonal types with no predominant pattern were identified. In addition, most isolates (71.4%) were VIM producers, suggesting that the genes encoding this type of carbapenemase may be transmitted horizontally between strains of this species.
It is important to be familiar with the drug resistance pattern in order to optimise empirical antimicrobial therapy in these patients. Our study shows that meropenem is the most active in vitro carbapenem in all isolated species and for all types of carbapenemase. It also reveals the high rates of resistance to aminoglycosides, fluoroquinolones and cotrimoxazole and especially to colistin (25%) and tigecycline (34.6%), two antibiotics used almost exclusively as rescue treatment for infection by these bacteria. A study conducted in 83 Spanish hospitals in 2013 found that among the CPE strains characterised, 4.5% were resistant to colistin and 29% to tigecycline.2 It is striking to note the growing colistin resistance in K. pneumoniae observed in our hospital, which has increased from 2.1% in 2011–20145 to 27.4% in 2015–2016. This is a sign of the increased resistance to this antibiotic in this species.
Our results also provide useful information regarding the possible use of new antibiotics, such as ceftazidime–avibactam in the treatment of CPE infections. This combination may play an important role in the treatment of some of these bacteria, since it is potentially active against OXA-48- and KPC-producing strains,10 although in vitro sensitivity to this new antimicrobial must be confirmed in each case.
This study has certain limitations. First, the microdilution panels used to perform the CPE antibiogram contained only imipenem and ertapenem, the latter at concentrations higher than those recommended for suspected carbapenemase production, so some CPEs might not have been identified. In addition, only OXA-48, VIM and KPC PCR were performed, as these are the most common carbapenemases in Spain. Finally, the K. pneumoniae PFGE study did not include all the isolates, only a representative sample, so it is likely that not all circulating clones in our hospital were detected.
Despite this, the study raises awareness of the serious problem of multi-resistant microorganisms. CPE infections have spread beyond the limits of our departments and the hospital itself to different types of patients and clinical settings. While we await the development of new antibiotics, it is important to review and intensify control measures and to update policies designed to encourage prudent use of antibiotics in hospitals and other care settings.
Conflicts of interestThe authors declare that they have no conflicts of interest.
Please cite this article as: Brañas P, Gil M, Villa J, Orellana MÁ, Chaves F. Epidemiología molecular de las infecciones/colonizaciones por enterobacterias productoras de carbapenemasas en un hospital de Madrid. Enferm Infecc Microbiol Clin. 2018;36:100–103.