Rapid diagnostic techniques are valuable tools in the diagnosis of gastrointestinal infections, especially for the detection of some microorganisms and in certain groups of patients. While antigen detection techniques are widely used in Clinical Microbiology laboratories, for the diagnosis of viruses, some parasites and some bacteria, molecular techniques are routinely used only for some pathogens (such as Clostridium difficile). However, molecular techniques are constantly evolving, and they allow a rapid diagnosis for an increasing number of pathogens, with high sensitivity and specificity. In addition, they are also able to detect virulence factors or resistance mechanisms. Syndromic surveillance systems, which detect different pathogens simultaneously, are very promising because they enable the most frequent pathogens to be diagnosed in a few hours and they can be very useful in certain patients.
For the diagnosis of Helicobacter pylori infection, molecular techniques are able to detect bacteria and its resistance to clarithromycin and levofloxacin, allowing the most appropriate treatment to be selected for each patient when bacterial culture is not possible.
Las técnicas de diagnóstico rápido son herramientas de gran valor en el diagnóstico de las infecciones gastrointestinales, especialmente para la detección de algunos microorganismos y en determinados grupos de pacientes. Mientras que las técnicas de detección de antígeno son práctica habitual en los laboratorios de microbiología clínica para el diagnóstico de virus, algunos parásitos y algunas bacterias, las técnicas moleculares se utilizan de manera rutinaria solo para determinados patógenos (como Clostridium difficile). Sin embargo, son técnicas en constante evolución que permiten el diagnóstico rápido de un número cada vez mayor de patógenos con una elevada sensibilidad y especificidad y también permiten la detección de factores de virulencia o mecanismos de resistencia. Los sistemas de diagnóstico sindrómico, que detectan diferentes patógenos de forma simultánea, son muy prometedores porque permiten diagnosticar en pocas horas los patógenos más frecuentes y pueden ser de gran utilidad en determinados pacientes.
Para el diagnóstico de la infección por Helicobacter pylori las técnicas moleculares, que pueden detectar tanto la bacteria como su resistencia a claritromicina y levofloxacino, permiten seleccionar el tratamiento más adecuado para cada paciente cuando el cultivo de la bacteria no es posible.
Infections of the gastrointestinal tract include 2 distinct clinical entities: gastric infections caused by Helicobacter pylori, and gastroenteritis, inflammation of the gastric and intestinal mucosa, which may be caused by various pathogens (bacteria, viruses, or parasites) or their toxins, and which includes diarrhoea, vomiting, abdominal pain and/or fever. Different pathogens may give rise to similar symptoms, making the aetiological diagnosis important. They are often self-limiting processes, but may present high morbidity and mortality rates, especially in young children or immunosuppressed patients.1 In developing countries they are associated with poor sanitation, lack of running water and nutritional deficiencies in the population, and present higher morbidity and mortality rates than in developed countries, where they are frequently associated with food-borne infections.2,3
The rapid and successful diagnosis of the pathogen involved in the clinical picture is important since some pathogens require a specific treatment; with some it is necessary to use control measures to avoid dissemination, and in some cases knowing the pathogen will enable a decision to be taken as to whether to hospitalise the patient or treat them on an outpatient basis.1,4
Types of rapid techniquesConventional diagnostic methods used in the diagnosis of gastroenteritis include bacterial culture techniques, which require several days to obtain results, microscopic tests for detection of eggs and parasites, requiring high specialisation of personnel, and antigen detection techniques.1 Frequently, when the aetiologic agent causing the clinical picture is successfully cultivated, the symptoms have already been resolved and it is not useful for the management of the patient.
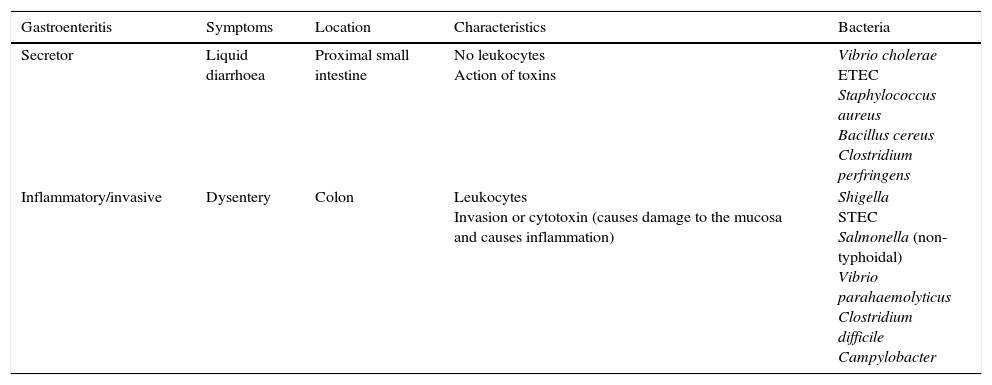
Rapid techniques based on microscopic observationGram staining, a technique used in most clinical specimens, is not useful in faecal samples, as they will stain the usual microbiota bacteria. It does allow for observation of the characteristic curved structures of Campylobacter, which may have value in certain circumstances, although it is not performed routinely.2 Microscopic examination for leukocytes is non-specific, but may be helpful if invasive enteritis is suspected (Table 1).
Types of diarrhoea in terms of mechanism of action.
| Gastroenteritis | Symptoms | Location | Characteristics | Bacteria |
|---|---|---|---|---|
| Secretor | Liquid diarrhoea | Proximal small intestine | No leukocytes Action of toxins | Vibrio cholerae ETEC Staphylococcus aureus Bacillus cereus Clostridium perfringens |
| Inflammatory/invasive | Dysentery | Colon | Leukocytes Invasion or cytotoxin (causes damage to the mucosa and causes inflammation) | Shigella STEC Salmonella (non-typhoidal) Vibrio parahaemolyticus Clostridium difficile Campylobacter |
The microbial antigens present in the biological samples can be detected and quantified by immunological techniques based on the specificity of the antigen-antibody reactions. A wide variety of immunoassays are available for the diagnosis of gastrointestinal tract infections. Some, such as immunofluorescence or enzyme-linked immunosorbent assay (ELISA), are laborious and require trained personnel, while others, such as membrane EIAs, lateral flow immunoassays or immunochromatographic tests (LFIA/ICT), and agglutination techniques, are extremely simple and fast.
ELISA enzyme-linked immunosorbant assayThey use specific antibodies for the test antigen, generally bonded to the wells of a microplate. The antigen present in the clinical sample is combined with the antibody bound to the solid phase and its presence is detected by the addition of a second antibody conjugated to an enzyme. The most frequently used enzymes are peroxidase and alkaline phosphatase. When the enzyme acts on the substrate, a colorimetric signal will be produced that is visible to the naked eye or quantifiable through the use of a spectrophotometer. In general, the sensitivity and specificity values of these techniques are excellent. They allow simultaneous processing of batches of samples and, in some cases, a quantification of the detected antigen. They are laborious techniques as they require the addition of multiple reagents, wash steps and incubation times. However, they are easily automated.
Membrane enzyme immunoassayThe capture antibody is immobilised in a membrane through which the sample flows. If the antigen is present, it will be retained by said antibody, and the antigen-antibody binding will become apparent, as in a conventional ELISA, through a second antibody conjugated to an enzyme. The interaction of the enzyme and its substrate causes a colour change. A determination is considered positive when coloured bands are obtained both in the control line (where the conjugate is attached) and in the reaction line (where the antigen is attached). Through this type of assay, the samples are processed individually, no equipment or specialised personnel are required. The results are obtained approximately in 15–20min, and are easily interpreted.
Immunochromatographic tests or lateral flow immunoassayThey consist of nitrocellulose or nylon membranes, like a strip or cassette, through which the sample flows by capillary action. In the most common sandwich assays, two zones are distinguished on the strip: the reaction zone, where antibodies against the test antigen are immobilised, and the control zone, where anti-conjugated antibodies are immobilised. The conjugate is an antibody, specific to the test antigen, labelled with a colloidal gold molecule or with coloured polystyrene microspheres. If the antigen is present in the sample, it will bind to both the conjugated antibody and the capture antibody immobilised in the reaction zone. The excess conjugated antibody will continue to migrate through the membrane until it is retained in the control zone. In negative samples, only the control zone will be coloured, whereas if the sample is positive, both the control and reaction zones will be coloured.5 These are qualitative techniques, very fast and simple to perform, which do not require special laboratory equipment and the samples are processed individually. Most present high values of sensitivity and specificity, although generally lower than those presented by conventional ELISA techniques.
Latex agglutination techniquesThey are based on the use of latex particles bound to the crystallisable fragment (Fc) of immunoglobulins. The antigen-binding fragments (Fab) are exposed and capable of binding to the antigen found in the sample. When the antibodies bind to the antigen, a lattice of the latex particles is produced which results in visible agglutination. These types of techniques require the specimens to be processed individually, are easy to perform, fast and do not require special equipment. However, cross reactions may occur with other sample antigens or prozone phenomena, in which an excess of antibodies prevents the formation of the framework.
Direct immunofluorescenceIt is based on the detection of antigens by the use of fluorochrome-labelled antibodies on extensions of clinical samples prepared on a slide. The extension is fixed, incubated with the antibody specific to the fluorochrome-conjugated test antigen and, after washing, the results are read by fluorescence microscopy. Its main disadvantages are the cost of the reagents and the laboriousness of the technique.
Rapid molecular techniquesMolecular techniques have revolutionised microbiological diagnosis and, despite their high cost, represent an interesting alternative to conventional methods due to their speed and high sensitivity and specificity. Polymerase chain reaction (PCR) reproduces the physiological process of DNA duplication in cells in vitro, exponentially amplifying a specific sequence of double-stranded DNA.
Since its invention, different variants of PCR have been designed that improved its diagnostic yield.
PCR after reverse transcription or RT-PCR is used for the detection and amplification of RNA. The RNA present in the sample is used as a template for synthesising the complementary DNA (cDNA) by inverse transcriptase or reverse transcriptase, and the resulting DNA is amplified by standard PCR.
Quantitative PCR (qPCR) or real-time PCR is much faster than the conventional version, and provides a continuous and precise quantification of the DNA that is being formed.6 In order to quantify the DNA or RNA present in the sample, it is necessary to perform a standard parallel curve.
Multiplex PCRs enable, through the use of multiple probes or pairs of primers, the simultaneous detection of different pathogens. These systems allow a syndromic diagnosis of gastrointestinal disease by simultaneously detecting the most frequently involved enteropathogens, including bacteria, viruses and parasites.
Other amplification techniques are available, such as isothermal amplification, in which nucleic acid amplification takes place at a constant temperature with the involvement of different enzymes such as reverse transcriptase, RNAse H, helicase or RNA polymerase.
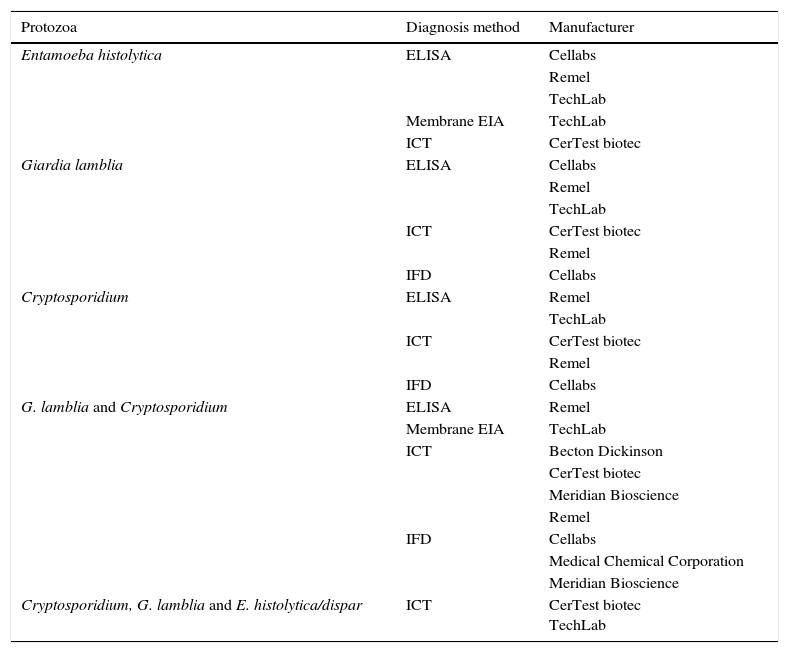
Rapid techniques for the detection of individual pathogensDiagnosis of parasitesTraditionally, the microbiological diagnosis of both protozoa (amoebas, flagellates, ciliates, coccidia and microsporidia) and helminths (nematodes, cestodes and trematodes) has been performed by microscopic identification of the parasitic forms that are expelled in the patient's faeces. To increase the diagnostic yield, it is necessary to use concentration techniques and, in some cases, specific stains. These determinations only permit the diagnosis of the acute and patent infection, require the taking of serial samples, are very laborious, require specialised personnel and present limitations in terms of sensitivity and specificity. Screening tests for parasitic antigens in faeces use specific antibodies that recognise secretion, surface, or wall molecules of parasites.7 There are commercialised systems that allow for the simultaneous detection of 2 or 3 different species (Table 2). The amplification tests of genomic sequences are those of greater sensitivity and specificity, and thanks to the constant characterisation of genomes of different organisms, it is possible to design PCR protocols for each of the parasites of interest. However, by using these techniques only one or a few parasites can be detected at a time and there are very few marketed.8
Commercialised assays, based on the detection of coproantigens, for the diagnosis of gastrointestinal tract infections caused by protozoa.
| Protozoa | Diagnosis method | Manufacturer |
|---|---|---|
| Entamoeba histolytica | ELISA | Cellabs |
| Remel | ||
| TechLab | ||
| Membrane EIA | TechLab | |
| ICT | CerTest biotec | |
| Giardia lamblia | ELISA | Cellabs |
| Remel | ||
| TechLab | ||
| ICT | CerTest biotec | |
| Remel | ||
| IFD | Cellabs | |
| Cryptosporidium | ELISA | Remel |
| TechLab | ||
| ICT | CerTest biotec | |
| Remel | ||
| IFD | Cellabs | |
| G. lamblia and Cryptosporidium | ELISA | Remel |
| Membrane EIA | TechLab | |
| ICT | Becton Dickinson | |
| CerTest biotec | ||
| Meridian Bioscience | ||
| Remel | ||
| IFD | Cellabs | |
| Medical Chemical Corporation | ||
| Meridian Bioscience | ||
| Cryptosporidium, G. lamblia and E. histolytica/dispar | ICT | CerTest biotec TechLab |
Entamoeba histolytica causes amoebic dysentery and is one of the possible causes of traveller's diarrhoea. The traditional diagnosis of amoebiasis has been based for many years on microscopy. However, microscopic techniques do not differentiate E. histolytica from the commensal amoebae Entamoeba dispar and Entamoeba moshkovskii. PCR techniques, based mostly on the amplification of coding regions of the small 18S ribosomal subunit (18S rRNA), differentiate the 3 species of morphologically indistinguishable Entamoeba and present excellent levels of sensitivity and specificity.9 On the other hand, the detection of E. histolytica antigens by ELISA is a good option if molecular methods are not available. Currently, different membrane EIAs and marketed ICT assays are available that detect E. histolytica-specific antigens in faeces and in abscess content.10,11 These assays are based on the detection of specific epitopes of amoebic lectin and have a sensitivity higher than that of microscopic examination.10,12 Some of the marketed assays only allow recent or frozen stools to be used.
Giardia lambliaGiardiasis, caused by Giardia lamblia, is a parasitic disease of great epidemiological and clinical importance due to its high prevalence and pathogenicity, mainly among children. Various methods of antigen detection with sensitivity and specificity levels superior to those of microscopic examination have been marketed for diagnosis. These tests are also useful in screening the infant population and as a treatment control to confirm healing. The various EIAs marketed use monoclonal or polyclonal antibodies to detect G. lamblia-membrane antigens such as GSA 65 or CWP1.11 They are rapid techniques, which are cost-effective and allow for the processing of recent stools or faeces preserved in formalin. For G. lamblia commercially available ICT assays are also available. However, most require using fresh or frozen stools, without preservatives, and have a lower sensitivity than ELISA assays. Direct immunofluorescence (DFI) stains for G. lamblia using a GSA 65 antigen-specific monoclonal antibody are also available.
CoccidiaCryptosporidium hominis and Cryptosporidium parvum are the 2 main pathogenic species of human parasite. Fundamentally they affect immunosuppressed patients and cause severe chronic diarrhoea. Coccidiosis is traditionally detected through the microscopic demonstration of oocysts following special stains such as Kinyoun or auramine-rhodamine. There are various other antigen detection techniques that are valid alternatives, such as IFD, ELISA or ICT techniques, that have shown specificity and sensitivity values higher than those of microscopic observation.7,11,13 However, none of the methods mentioned allows the identification of species, so it is necessary to resort to PCR techniques.
HelminthsThere are 3 groups of helminths of medical importance: nematodes (roundworms), cestodes (bandworms) and trematodes (or staves). The diagnosis of some cestodes is based on the observation of proglottids or segments. However, in most helminth infections, diagnosis is made by microscopic identification of their eggs or larvae in the patient's stool. Although there are also special techniques, valid only for certain helminths, such as the Graham ribbon in the diagnosis of Enterobius vermicularis or the cultivation of Strongyloides stercoralis, advances in the rapid diagnosis of intestinal helminths are not as significant as in protozoa. Antigen detection tests have not been highly developed and, although several studies have been published,14 they have not yet been marketed. More frequent are the serological tests of specific antibodies, available for the diagnosis of hydatidosis, cysticercosis, fasciolosis, schistosomiasis, toxocariasis, anisakiasis, filariasis and strongyloidiasis. Some of these assays have been marketed, while others are only carried out at reference centres. They all have the drawback of not distinguishing between current and past infection, so they are more useful in non-endemic areas, and often have cross-reactivity problems15,16 As for the molecular diagnosis, a multitude of PCR protocols have been designed, both conventional and real-time, for the diagnosis of helminthiasis.8 However, so far none has been marketed.
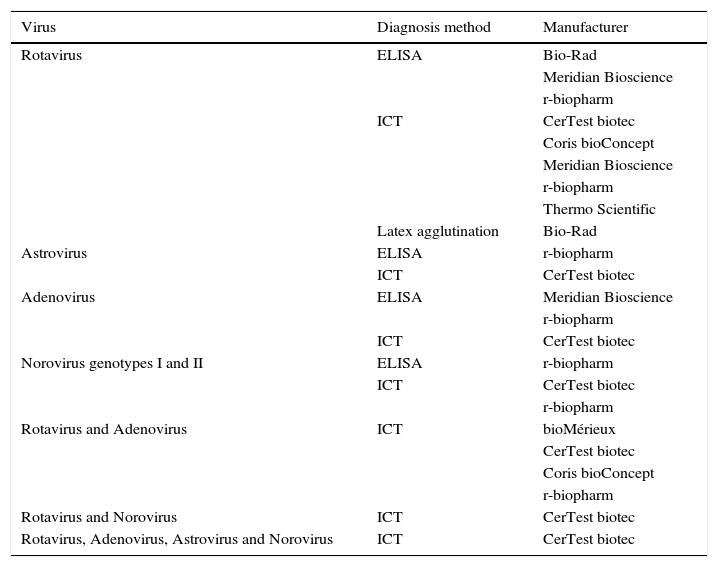
Diagnosis of virusThe main viruses producing gastroenteritis in humans are rotavirus, astrovirus, adenovirus and norovirus. Others such as coronaviruses, torovirus, picobirnaviruses and picornaviruses (Aichi virus) can also cause gastrointestinal tract infection, but with a much lower frequency.16 The rapid diagnosis of viral gastroenteritis is made through the detection of viral antigens (Table 3) or by molecular methods based on the detection of specific genes.
Commercialised assays, based on the detection of coproantigens, for the diagnosis of gastrointestinal tract infections caused by viruses.
| Virus | Diagnosis method | Manufacturer |
|---|---|---|
| Rotavirus | ELISA | Bio-Rad |
| Meridian Bioscience | ||
| r-biopharm | ||
| ICT | CerTest biotec | |
| Coris bioConcept | ||
| Meridian Bioscience | ||
| r-biopharm | ||
| Thermo Scientific | ||
| Latex agglutination | Bio-Rad | |
| Astrovirus | ELISA | r-biopharm |
| ICT | CerTest biotec | |
| Adenovirus | ELISA | Meridian Bioscience |
| r-biopharm | ||
| ICT | CerTest biotec | |
| Norovirus genotypes I and II | ELISA | r-biopharm |
| ICT | CerTest biotec | |
| r-biopharm | ||
| Rotavirus and Adenovirus | ICT | bioMérieux |
| CerTest biotec | ||
| Coris bioConcept | ||
| r-biopharm | ||
| Rotavirus and Norovirus | ICT | CerTest biotec |
| Rotavirus, Adenovirus, Astrovirus and Norovirus | ICT | CerTest biotec |
Rotaviruses are highly contagious RNA viruses that are transmitted by the faecal-oral route, by fomites or by the respiratory route. According to the antigenic characteristics of the VP6 glycoprotein, a component of the internal virion capsid, 8 serogroups (A-H) are distinguished. Human infections are mainly produced by rotavirus serogroup A and, to a lesser extent, by serogroups B and C. The VP4 protein, which constitutes the virion spicules, and the VP7 glycoprotein, constituent of the outer capsid, determine the existence of the serotypes P and G, respectively.17 The diagnosis of serogroup A rotavirus infections is based on the detection of viral antigens in stool samples, specifically the VP6 glycoprotein. At the moment there are no commercially available methods to detect rotavirus from other serogroups. The formats available are highly varied: ELISA, membrane EIA, ICT and latex agglutination.18–20 The membrane EIA, the ICT techniques and latex agglutination are suitable methods for hospitals with few or sporadic samples. However, they are less sensitive than conventional ELISAs. The more sensitive and specific molecular methods are based on an RT-PCR. For the diagnosis of group A rotavirus by RT-PCR, marketed systems use specific primers of the gene encoding the NSP3 non-structural protein. This methodology also allows the detection of rotavirus of different serogroups.21
AstrovirusAstroviruses are non-enveloped viruses with a positive-sense single-stranded RNA genome, small (28–41nm in diameter) and star-shaped under an electron microscope. There are 8 different serotypes (1–8) but, due to the existence of antigenic cross reactions, it is possible to detect all of them using a group-specific antibody called MAb8E7, capable of recognising an epitope common to all human serotypes. Several commercial ELISA kits are currently available to allow the detection of human astrovirus in faeces. The diagnosis can also be established by RT-PCR.22
AdenovirusEnteric adenoviruses of serotypes 40 and 41 are the most frequently associated with childhood gastroenteritis. There are specific type methods that detect only serotypes 40 and 41 and other specific group methods that detect all adenoviruses. The diagnosis of enteric adenovirus infections is usually established by ELISA, latex agglutination or ICT. Some of these techniques allow simultaneous detection of adenovirus and rotavirus. There are also commercially available highly sensitive and specific PCR methods that allow the detection and characterisation of enteric adenoviruses.16
NorovirusNoroviruses are the leading cause of non-bacterial gastroenteritis in individuals of all ages and are the most common cause of outbreaks of acute gastroenteritis. They are non-enveloped viruses, with a diameter of approximately 38nm, whose genome is made up of positive-sense single-stranded RNA. Of the 5 genogroups described, only genogroups I, II and IV have strains that infect humans. RT-PCR has become the reference technique for its diagnosis. The primers employed enable amplification of the viral RNA polymerase gene or the gene encoding the capsid proteins. Real-time RT-PCR is more sensitive and allows to detect and differentiate between genogroups I and II more quickly. In addition, there are several diagnostic tests based on the detection of antigens.23 Some ELISAs have a high specificity but a moderate sensitivity, so they are very useful in the study of epidemic outbreaks, but not so much in the diagnosis of sporadic cases. However, the available ICTs and membrane EIAs have high sensitivity and specificity values.
Diagnosis of bacteriaToxigenic Clostridium difficileClostridium difficile is a spore-producing anaerobic gram-positive bacillus found in the soil and the intestinal tracts of animals and humans and can colonise up to 50% of children under one year old, 3–5% of healthy adults, and a higher percentage of hospitalised adult patients or those in nursing homes. C. difficile produces antibiotic-associated diarrhoea, but symptoms may range from asymptomatic carriers, to moderate or mild diarrhoea and pseudomembranous colitis.2,24,25 Pathogenic strains have a pathogenicity locus that codes for toxin A, an enterotoxin encoded by the TcdA gene, and toxin B, a potent cytotoxin encoded by the TcdB2,25 gene, although they may not present toxin A and only toxin B. The hypervirulent strain NAP1/027/B1 has been associated with increased morbidity and mortality rates and produces greater toxin expression, more efficient sporulation and expression of a binary toxin, although not all the strains currently detected have these virulence characteristics.2,26
In patients with risk factors for C. difficile diarrhoea, both hospitalised and non-hospitalised, rapid detection of toxins A and B should be performed to initiate antibiotic treatment and to apply control measures to prevent it spreading to other patients.
The most commonly used methods are:
Antigen detection. Detection of glutamate dehydrogenase (GDH) by EIA or latex particles. GDH is a very abundant antigen of the cell wall of C. difficile and its detection is very sensitive but not very specific, since it is present in both toxigenic and non-toxigenic strains.2,24,27
Detection of toxins A and/or B. It can be done by cytotoxicity tests or by ICT or EIA techniques.4,28 The cytotoxicity test is considered the reference method for detection of toxin B in the faeces, with a high sensitivity and specificity, but it is very laborious, in addition to not being a rapid test. There are numerous commercial EIA tests for the detection of toxins A, B or both. It is recommended to use EIA techniques that detect toxin B or both, because of the producing strains of toxin B. However, these commercial techniques have low sensitivity compared to cell cultures, so nucleic acid detection techniques must be used.2,28 The detection of toxin from the isolation obtained by culture should not be used as a single method, since it requires several days to grow, but its use, in addition to the direct detection of toxin, can increase the positivity by 10%.29
Detection of toxin A/B genes by nucleic acid amplification. It is the technique that offers greatest sensitivity and specificity, although it is more expensive. There are different real-time PCRs that allow detection of C. difficile, toxin B, and binary toxin in 90min. GeneXpert presents a 98% level of accordance with the reference test and greater sensitivity than the ICT.4,30 However, a sample can be positive by molecular method and negative by EIA and cell culture, and as a result doubt can arise from the clinical meaning.2,27
To arrive at a correct diagnosis and to be cost-effective, one of the following algorithms must be used2,24,25: (1) perform a GDH EIA and study the presence of toxins using the molecular method when positive, (2) perform a combination of GDH and toxin A/B EIA simultaneously and confirm the samples showing discrepant results using the molecular method or (3) perform only the molecular method on all samples.
Escherichia coliEscherichia coli may be part of the normal intestinal microbiota, but some variants (considered pathovarieties or pathotypes) have the capacity to produce diarrhoea and are a cause of morbidity and mortality around the world.31 They are defined by the expression of one or more virulence factors: enteropathogen (EPEC), Shiga toxin producer (STEC), also called enterohemorrhagic or verocytotoxin producer, which includes serotype O157, enteroinvasive (EIEC), enterotoxigenic (ETEC) enteroaggregative (EAEC) and adherent-invasive (AIEC).2,31
In most clinical and public health laboratories only STEC is studied and is defined by the presence of Shiga toxin 1 (stx1) and/or Shiga toxin 2 (stx2) genes. The culture allows only E. coli O157 to be detected, but culture-independent methods detect both O157 and the others.2 The CDC recommends simultaneously preparing the differential culture to detect E. coli O157 and the molecular study for the detection of stx1 and stx2 in faeces of patients with acute diarrhoea acquired in the community and with possible haemolytic uraemic syndrome.32 If a molecular method is not available, it should be sent to a reference centre.
The following can be used: (1) EIA Stx, which can detect and in some cases differentiate between stx1 and stx2, from an enriched broth incubated overnight at 37°C, of both STEC O157 and non-O157, and (2) molecular techniques, which detect the stx1 and stx2 genes directly from faeces, and which are the most sensitive method.2 Recently the use of a new ICT performed directly on stool samples with excellent results has been published.30
CampylobacterCampylobacter antigen detection tests have been developed from stool samples, in ELISA format, on a microwell plate or in lateral flow ICT format. Although they are faster methods than the culture, they are less sensitive and do not differentiate between Campylobacter jejuni and Campylobacter coli. There are molecular methods that can be performed directly from faeces, which are the most sensitive, but are not sufficiently validated.2
Rapid techniques for detection of other diarrhoea-producing bacteriaA rapid test (ICT, dipstick) detects Shigella from colonies, and directly from faeces or rectal exudates, thanks to monoclonal antibodies against lipopolysaccharide.4 The enterotoxins of Staphylococcus aureus, Bacillus cereus and Clostridium perfringens can be detected in food, faeces or in strains cultured by commercial methods, by means of the reversed passive latex agglutination technique or EIA technique.33
Helicobacter pyloriFor the rapid diagnosis of H. pylori infection, different techniques have been used, since the culture of the biopsy is slow and demanding.34
Based on H. pylori urease. H. pylori has a potent urease and this characteristic has been used, since the discovery of H. pylori, for its diagnosis, in 2 types of assays: rapid urease from the biopsy and breath assay.
If a piece of biopsy tissue is incorporated into a urea broth or gel, the colour will change to pink in a few minutes. The urease test can be used to identify the bacteria obtained by culture, but it is mainly used as a standard practice in endoscopy units. The first to be marketed for this purpose was the CLO-test, with which positive results were obtained between 30min and 3h, although it could be left up to 24h, which led to a loss of specificity due to other urease positive bacteria. Currently there are different trademarks, which seek to shorten the time to obtain the result.35
To perform the urea breath test (UBT), the patient ingests marked urea and breath samples are collected before and 30min after ingestion. If H. pylori is in the stomach, it will hydrolyse the urea and marked CO2 will be released. A radioactive isotope can be used, 14C, or a non-radioactive isotope, 13C, which is the most widely used and can be measured with a mass or infrared spectrometer.35 It is the non-invasive test of choice for the diagnosis and follow-up of patients with H. pylori because of their high sensitivity and specificity (95–100%) to detect active infection.35,36
Based on the detection of antibodies. There are numerous serological tests to detect anti-H. pylori IgG antibodies in the patient's serum. The main limitation is that they do not differentiate the active infection from the past infection and the kit used needs to be validated locally.35 It is useful for epidemiological studies and when there is gastrointestinal bleeding, atrophic gastritis, gastric cancer or MALT lymphoma (mucosa associated lymphoid tissue), since it can produce a low bacterial load in the stomach and will reduce the sensitivity of other diagnostic tests.36,37 There are also tests to detect H. pylori antibodies in urine or saliva, with variable results, so it is currently not recommended for use in patient management.35,38,39
Based on the detection of antigens in faeces. The stool antigen test is a non-invasive and qualitative test that detects the H. pylori antigen that has been eliminated in faeces. They can be used: (1) as a test for the initial diagnosis of infection; (2) for treatment control (at least 4 weeks after finishing treatment), and (3) for epidemiological studies.
The first kit to be developed was an EIA with polyclonal antibodies,39 although it is currently not recommended because of its lower sensitivity and specificity compared to those that use monoclonal antibodies.35,39 Of the numerous methods marketed that use monoclonal antibodies, EIA or ICT present different diagnostic precision. Better results are obtained with kits based on EIA in microwell format39 and are considered equivalent to the UBT in the Maastricht consensus report.34,35 There is also an automated chemiluminescent immunoassay with results comparable to the EIA.34
Sensitivity of 93% and 95% and specificity of 89% and 87% for the pretreatment diagnosis and sensitivity of 94% and 100% and specificity of 97% and 91% for post-treatment follow-up were observed in 2 studies evaluating ICT.35 In a recent study, 3 commercial ICTs were compared and RAPID Hp StAR (Oxoid) was found to be the most sensitive (91–92%) but less specific (77–85%), whereas for Uni-Gold™ H.pylori Antigen (Trinity Biotech) and for ImmunoCard STAT! HpSA (Meridian), a sensitivity of 83% and a specificity of 90% (87–89%) were observed.40 In other studies, lower values of sensitivity and/or specificity have also been observed, therefore it is believed that ICTs need to be improved to be reliable.40
Based on molecular methods (PCR). For many years, home molecular methods have been used for the diagnosis of H. pylori infection from different samples such as gastric biopsy, faeces, saliva, gastric juice, etc. In addition, they allow for the study of macrolide resistance mutations such as clarithromycin (23S rRNA gene), fluoroquinolones (gyrA gene), tetracyclines (16S rRNA gene), rifabutin (rpoB gene) and amoxicillin (pbp-1a gene),41,42 and have the advantages of speed, greater sensitivity than culture with phenotypic methods of antibiotic sensitivity, and they detect heterogeneous resistance, which may be difficult to obtain by culture.39,43
In the last Maastricht consensus, sensitivity studies on clarithromycin are considered if this antibiotic is to be used as the first line of treatment in regions with a resistance rate greater than 15%, both by culture and antibiogram, and by a molecular method directly from the biopsy.36
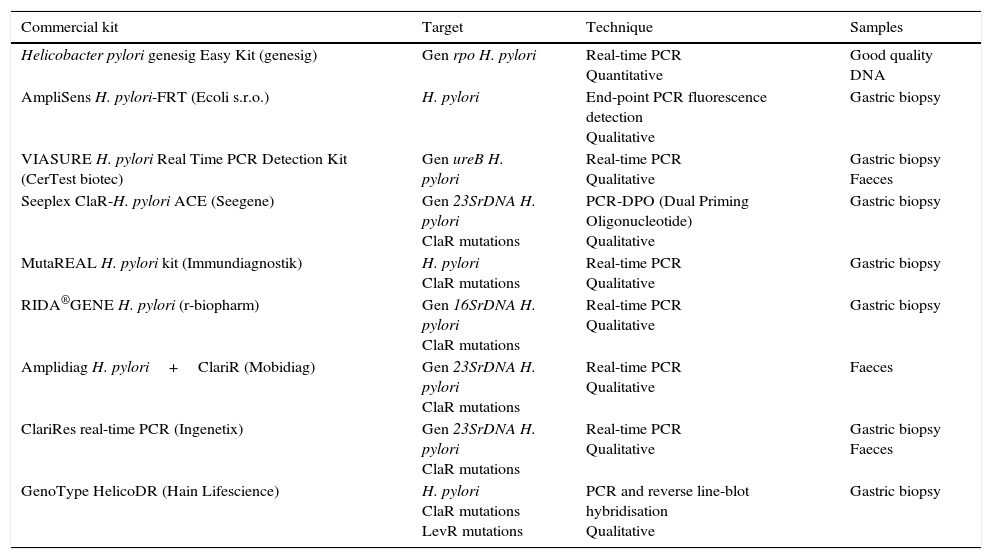
There are several commercial kits based on real-time PCR that detect H. pylori directly on the biopsy; other methods, in addition to H. pylori, detect the major mutations conferring resistance to clarithromycin44–47 (Table 4). There is also a commercial kit based on PCR followed by reverse line-blot hybridisation (GenoType HelicoDR) which detects resistance to clarithromycin and fluoroquinolones. Sensitivity of 94% to 100% and specificity of 86% to 99% have been reported for the detection of clarithromycin resistance and sensitivity of 83% to 87% and specificity of 95% to 98.5% for fluoroquinolone resistance,39,48 although in other studies lower levels have been observed for both clarithromycin and fluoroquinolones.39,49 Some of the commercialised kits can detect H. pylori and its resistance to clarithromycin from stool samples (Amplidiag H. pylori+ClariR, Mobidiag or ClariRes real-time PCR, Ingenetix), although no articles have yet beem published.
Commercial systems using molecular methods for the diagnosis of Helicobacter pylori and resistance to clarithromycin or levofloxacin.
| Commercial kit | Target | Technique | Samples |
|---|---|---|---|
| Helicobacter pylori genesig Easy Kit (genesig) | Gen rpo H. pylori | Real-time PCR Quantitative | Good quality DNA |
| AmpliSens H. pylori-FRT (Ecoli s.r.o.) | H. pylori | End-point PCR fluorescence detection Qualitative | Gastric biopsy |
| VIASURE H. pylori Real Time PCR Detection Kit (CerTest biotec) | Gen ureB H. pylori | Real-time PCR Qualitative | Gastric biopsy Faeces |
| Seeplex ClaR-H. pylori ACE (Seegene) | Gen 23SrDNA H. pylori ClaR mutations | PCR-DPO (Dual Priming Oligonucleotide) Qualitative | Gastric biopsy |
| MutaREAL H. pylori kit (Immundiagnostik) | H. pylori ClaR mutations | Real-time PCR Qualitative | Gastric biopsy |
| RIDA®GENE H. pylori (r-biopharm) | Gen 16SrDNA H. pylori ClaR mutations | Real-time PCR Qualitative | Gastric biopsy |
| Amplidiag H. pylori+ClariR (Mobidiag) | Gen 23SrDNA H. pylori ClaR mutations | Real-time PCR Qualitative | Faeces |
| ClariRes real-time PCR (Ingenetix) | Gen 23SrDNA H. pylori ClaR mutations | Real-time PCR Qualitative | Gastric biopsy Faeces |
| GenoType HelicoDR (Hain Lifescience) | H. pylori ClaR mutations LevR mutations | PCR and reverse line-blot hybridisation Qualitative | Gastric biopsy |
ClaR: resistance to clarithromycin; LevR: resistance to levofloxacin.
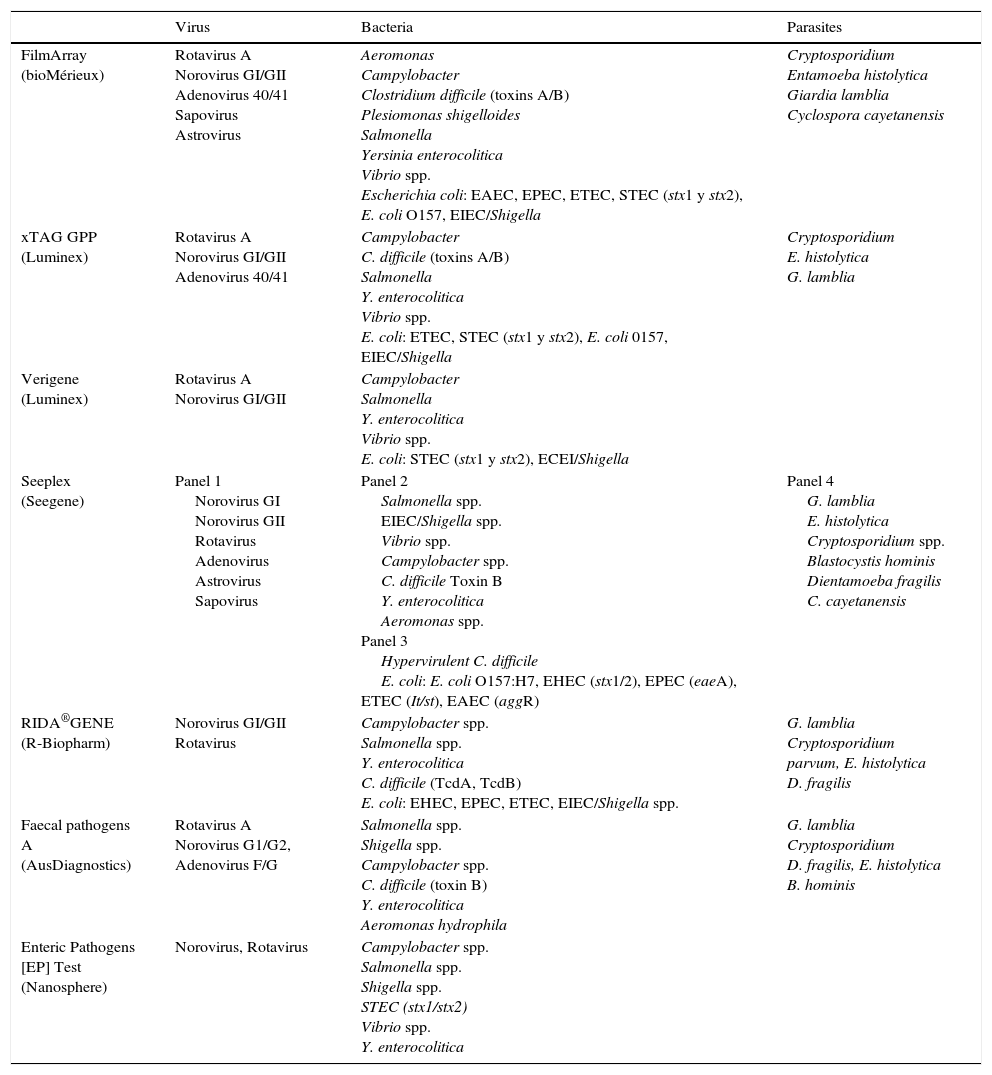
Numerous syndromic panels have emerged in recent years for the microbiological diagnosis of different infectious diseases, including diarrhoea-producing pathogens. Within a short time period, they deliver results of all the pathogens included in the kit and can be a very important tool in some situations, such as in immunosuppressed patients or serious patients.1 The detection of most pathogens has a good sensitivity and it is highly valuable in the detection of coinfections.4
There are different commercial techniques to detect gastroenteritis-producing pathogens (Table 5), and they vary widely in the number and type of pathogens they detect and in the technology they use.1,2,50
Pathogens detected in commercial panels for the diagnosis of gastroenteritis.
| Virus | Bacteria | Parasites | |
|---|---|---|---|
| FilmArray (bioMérieux) | Rotavirus A Norovirus GI/GII Adenovirus 40/41 Sapovirus Astrovirus | Aeromonas Campylobacter Clostridium difficile (toxins A/B) Plesiomonas shigelloides Salmonella Yersinia enterocolitica Vibrio spp. Escherichia coli: EAEC, EPEC, ETEC, STEC (stx1 y stx2), E. coli O157, EIEC/Shigella | Cryptosporidium Entamoeba histolytica Giardia lamblia Cyclospora cayetanensis |
| xTAG GPP (Luminex) | Rotavirus A Norovirus GI/GII Adenovirus 40/41 | Campylobacter C. difficile (toxins A/B) Salmonella Y. enterocolitica Vibrio spp. E. coli: ETEC, STEC (stx1 y stx2), E. coli 0157, EIEC/Shigella | Cryptosporidium E. histolytica G. lamblia |
| Verigene (Luminex) | Rotavirus A Norovirus GI/GII | Campylobacter Salmonella Y. enterocolitica Vibrio spp. E. coli: STEC (stx1 y stx2), ECEI/Shigella | |
| Seeplex (Seegene) | Panel 1 Norovirus GI Norovirus GII Rotavirus Adenovirus Astrovirus Sapovirus | Panel 2 Salmonella spp. EIEC/Shigella spp. Vibrio spp. Campylobacter spp. C. difficile Toxin B Y. enterocolitica Aeromonas spp. Panel 3 Hypervirulent C. difficile E. coli: E. coli O157:H7, EHEC (stx1/2), EPEC (eaeA), ETEC (It/st), EAEC (aggR) | Panel 4 G. lamblia E. histolytica Cryptosporidium spp. Blastocystis hominis Dientamoeba fragilis C. cayetanensis |
| RIDA®GENE (R-Biopharm) | Norovirus GI/GII Rotavirus | Campylobacter spp. Salmonella spp. Y. enterocolitica C. difficile (TcdA, TcdB) E. coli: EHEC, EPEC, ETEC, EIEC/Shigella spp. | G. lamblia Cryptosporidium parvum, E. histolytica D. fragilis |
| Faecal pathogens A (AusDiagnostics) | Rotavirus A Norovirus G1/G2, Adenovirus F/G | Salmonella spp. Shigella spp. Campylobacter spp. C. difficile (toxin B) Y. enterocolitica Aeromonas hydrophila | G. lamblia Cryptosporidium D. fragilis, E. histolytica B. hominis |
| Enteric Pathogens [EP] Test (Nanosphere) | Norovirus, Rotavirus | Campylobacter spp. Salmonella spp. Shigella spp. STEC (stx1/stx2) Vibrio spp. Y. enterocolitica |
Luminex xTAG GPP. It detects bacteria, viruses and parasites. FDA approved and with CE marking, 24 samples can be made in about 5h in a process that includes: pretreatment, nucleic acid extraction, multiplex PCR, pellet hybridisation, and data analysis. This technique increases the rate of positivity with respect to the conventional methods, and detects mixed infections.1,51,52
FilmArray GI panel (bioMérieux). It detects bacteria, parasites and viruses. FDA approved and with EC marking, it consists of a closed device that requires minimal manipulation. The system performs nucleic acid extraction, nested multiplex PCR, and melting temperature analysis. Performing the test along with the analysis takes about 60min. Higher rates of positivity and a significant percentage of mixed infections are detected, more frequent in children under 5 years or in non-hospitalised patients. The most frequent pathogens in co-infection were Campylobacter and EPEC.1,53
Allplex™ Gastrointestinal Full Panel Assay (Seegene) is a system with 4 panels that detects viruses, bacteria and parasites and is approved for use in Europe (EC marked).
There are more commercial systems such as Nanosphere Verigene (Luminex), Prodesse ProGastro SSCS (Hologic Gen-Probe), BD MAX enteric bacterial panel (BD), EntericBio real-time Gastro (Serosep), CLART EnteroBac and Enteric Pathogens [EP] Test (Nanosphere).1,2,50
Analysis of volatile organic acidsVolatile Organic Acids are a broad range of stable chemical compounds that are volatile at room temperature and can be detected in patients’ breath, urine, faeces, and sweat.54 It has proven useful in the diagnosis of different diseases such as diabetes, gastrointestinal and liver diseases, lung diseases, different types of cancer and in various infections, such as gastrointestinal infections and H. pylori infection.55–57
At least 40 volatile acids can be detected in stool samples from patients with gastroenteritis, regardless of clinical disease, and different markers are observed depending on the specific type of pathogen.54 In Bangladeshi patients with Vibrio cholerae infection, fewer volatile compounds are observed than in healthy donor samples.58 In a study of 35 patients with infectious diarrhoea, compared to 6 controls, there were no hydrocarbons and terpenes in patients with Campylobacter infection, and the absence of non-native furan species was indicative of infections with C. difficile.54
In the exhaled breath analysis of patients infected with H. pylori, 3 volatile compounds (isobutane, 2-butanone, and ethyl acetate) were observed that were not detected in the controls.54,59 These compounds can be produced by H. pylori in vivo or be metabolites that the host produces as a result of infection. Elevated levels of hydrogen cyanide (HCN) and hydrogen nitrate have also been reported in the exhaled breath of patients with H. pylori gastritis compared to healthy controls. However, HCN is not specific to H. pylori, as it can be detected in exhaled breath samples from patients infected with Pseudomonas aeruginosa.54
ConclusionsRapid techniques are extremely valuable in the diagnosis of gastrointestinal infections. Those based on the detection of antigens are already common practice in clinical microbiology laboratories, as are those based on molecular techniques, although only for some pathogens such as C. difficile and Shiga toxin-producing E. coli. When choosing the technique and trademark to use, we must consider different criteria, which will include: the pathogen or pathogens we want to detect, the type of patients for whom it is intended, the number of requests we think need to be made, the availability of equipment in the department, as well as the cost of the technique. If we are going to conduct a small number of tests, individual techniques such as EIA or ICT would be most appropriate, while if the number of tests is high, it will be more convenient to perform ELISA tests. Molecular techniques that detect different pathogens simultaneously can be very useful in certain groups of patients and will become increasingly important because of their high sensitivity and specificity, as well as their speed, albeit at a higher cost.
Conflicts of interestT.A.C. has received sporadic support for attendance at congresses of Hain Lifesciencie, bioMerieux, Meridian.
Please cite this article as: Balsalobre-Arenas L, Alarcón-Cavero T. Diagnóstico rápido de las infecciones del tracto gastrointestinal por parásitos, virus y bacterias. Enferm Infecc Microbiol Clin. 2017;35:367–376.