The role of Aeromonas species in gastrointestinal disease is controversial. The aim was to analyze not only the virulence genes between different species of Aeromonas isolated from feces, but the distribution of these virulence genes between enterotoxigenic strains and co-pathogen strains.
MethodsRetrospective study of isolates of Aeromonas spp. in feces (2016–2021). The protocol included coproculture, identification by MALDI-TOF and confirmation by multiplex PCR. SPSS Statistics program was used.
ResultsA total of 288 strains were studied for the virulence genes between different species of Aeromonas. To compare virulence genes between Aeromonas as co-pathogen and those isolated alone, 218 strains of the global set were used; 52 as co-pathogens compared with 166 Aeromonas without associated pathogen as controls.
ConclusionsWe found no significant differences in the distribution of virulence genes versus co-existence of co-pathogens or not. A. hydrophila is the potentially most virulent species of our set.
El papel de las especies de Aeromonas en las enfermedades gastrointestinales es controvertido. El objetivo fue analizar no solo los genes de virulencia entre diferentes especies de Aeromonas aisladas de heces, sino también la distribución de estos genes de virulencia entre cepas enterotoxigénicas y co-patógenas.
MétodosEstudio retrospectivo de aislamientos de Aeromonas spp. en heces (2016-2021). El protocolo incluyó coprocultivo, identificación por MALDI-TOF y confirmación por PCR multiplex. Se utilizó el programa SPSS Statistics.
ResultadosSe estudiaron un total de 288 cepas para los genes de virulencia entre diferentes especies de Aeromonas. Para comparar genes de virulencia entre Aeromonas como co-patógeno y los aislados únicos, se utilizaron 218 cepas del conjunto global; 52 como co-patógenos, comparados con 166 Aeromonas sin patógeno asociado como controles.
ConclusionesNo se encontraron diferencias significativas en la distribución de los genes de virulencia versus coexistencia de co-patógenos o no. Aeromonas hydrophila es la especie potencialmente más virulenta de nuestro muestreo.
Aeromonas species are aerobic and anaerobic Gram-negative rods, belongs to the Aeromonadaceae family and comprises a group of Gram-negative bacteria widely distributed in aquatic environments, but its great capacity for adaptation also makes Aeromonas species able to colonize terrestrial environments. Some clinically isolated Aeromonas spp. are pathogenic to humans.1 The genus Aeromonas comprises of two different groups of bacteria. One is non-motile psychrophilic Aeromonas salmonicida and the other group comprising of three mesophilic motile species: Aeromonas hydrophila, Aeromonas caviae, and Aeromonas sobria.2
Gastroenteritis is the most common type of Aeromonas infection in humans, but clinical presentation may include syndromes like skin and soft tissue infections, urinary tract infections, and central line associated bloodstream infections.3
Intestinal and extraintestinal infections by Aeromonas spp., remain controversial, due to the existence of healthy carriers of Aeromonas spp. In some studies, Aeromonas strains isolated from patients with enteritis was enterotoxigenic, while Aeromonas strains isolated from healthy people rarely showed enterotoxic characteristics forming part of the human gastrointestinal microbiota, varying the rates of frequency from 0% to 4% in people with no disease to 0.8%–7.4% in persons with diarrheal illness.4 The prevalence in case of diarrhea in pediatric age is around 4.6%5 and in other cases may even appear as a co-pathogen with other enteropathogen, especially in children, with a prevalence of 1.9%–57.7% in pediatric age.6,7 In our health area Aeromonas spp. is the third most frequently isolated enteropathogen in feces after Campylobacter spp. and Salmonella spp. without finding significant differences regarding the distribution by sex (52.0% women) or age, with a mean age of infection of 34 years. There are 15.8% of co-infections, being more frequent in pediatric age (49; 84.5%) (p=0.01) and mostly associated with Campylobacter spp. (75.9%).8
Under this context, the objective of this work was to identify the prevalence of five virulence genes in each species of Aeromonas studied, in this way we could advance the potentially most pathogenic species for humans. Also, the secondary objective of the work was to try to advance in clarifying which could be the potentially pathogenic species that affect humans, for this, we compared the presence of five virulence genes among those Aeromonas identified with another primary pathogen, considering as primary pathogen: Salmonella spp., Yersinia spp., Shigella spp. and Campylobacter spp., and Aeromonas isolated as a single pathogen in feces.
Methods288 isolates of Aeromonas spp. were studied, only one isolation per patient was considered, isolated in stool between 2016 and 2020 in the Microbiology Service of the University Hospital Marqués de Valdecilla, Santander (313.040 census population). The protocol for these isolates included inoculated on Yersinia Selective Agar (CIN Agar; BD, Heidelberg, Germany) and incubated at 37°C for 24h. Identification was performed by MALDI-TOF system (Vitek-MS®, BioMerieux) and were confirmed by multiplex PCR developed by Perssons et al.,9 and when there was not correlation between both systems, the rpoB gene sequencing was used. Also the presence of five virulence genes were studied by PCR: genes encoding heat-labile cytotonic enterotoxin (alt) and heat-stable cytotonic enterotoxin (ast), as well as genes encoding hemolysin (hlyA), aerolysin (aerA) and cytotoxic enterotoxin (act).10 For the isolation of copathogens, feces were seeded in MacConkey, XLD, CIN agar plates (Thermo Fisher Diagnostics, Hemel Hempstead, United Kingdom) and selective enrichment broth (Selenite Broth, BioMerieux) at 37°C for 24h. In Campylobacter selective agar plates (CCDA selective medium, Thermo Fisher Diagnostics, Hemel Hempstead, United Kingdom) in microaerophilia ambient at 37°C for 48h, following standard laboratory procedures. Identification was performed by MALDI-TOF system (Vitek-MS®, BioMerieux). Enteropathogenic viruses were also ruled out by immunochromatography (CerTest, Biotec, Zaragoza, Spain). On the other hand, it was compared whether there are significant differences in the presence of the different virulence genes with the coexistence or not of other gastrointestinal pathogens. For this, those strains that had been isolated as co-pathogenic were selected from our population, comparing them with a control group from the same population randomly matched by species of Aeromonas isolated, age and sex of the patient.
The SPSS Statistics program was used for statistical analysis. The statistical test used for the normality study was Shapiro Wilks, and for the intergroup comparison an ANOVA test was performed with subsequent post hoc analysis for a group of non-parametric Kruskal–Wallis samples. Significant results were considered for p-value <0.05. For stratification by age, adults aged 16 and over are considered.
ResultsThe distribution of the strains studied was as follows: A. caviae (n: 230; 82%) A. veronii (n: 26; 9%) A. hydrophila (n: 16; 5%) A. media (n: 15; 5%) A. salmonicida (n: 1; 0.3%), with a distribution in children of A. caviae (n: 112; 98.2%), A. veronii (n: 2; 1.8%). And on the other hand we have studied the virulence genes of 52 Aeromonas spp. isolated as co-pathogens against 166 Aeromonas spp. isolated without associated pathogen. The 52 Aeromonas spp., recovered with an associated primary pathogen, were isolated in feces from 8 (15.4%) adults and 44 (84.6%) were isolated from children. Most of them corresponded to the species A. caviae (47, 90.4%), while to a lesser extent they were A. veronii (4, 7.7%) and A. hydrophila (1, 1.9%). Also most of these Aeromonas spp. were isolated together with Campylobacter spp. (45, 86.6%) and lower percentage together with Salmonella spp. (6, 11.5%) and Yersinia spp. (1, 1.9%). Aeromonas spp. used as controls were isolated from patients with the same age ranges as the cases.
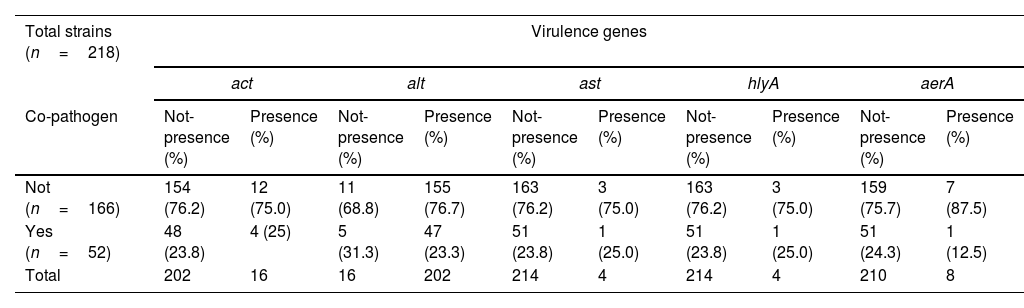
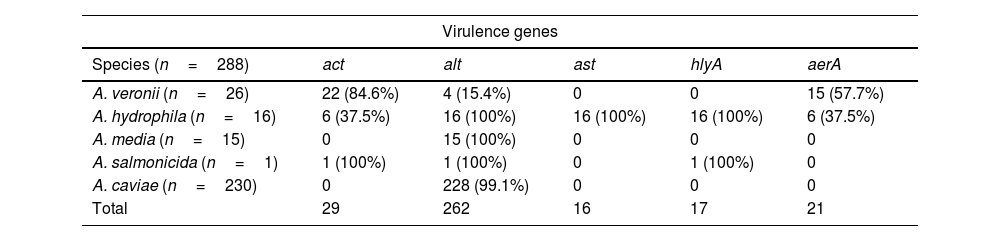
No significant differences were found in the distribution of virulence genes versus co-existence of co-pathogens with p value 0.911; 0.472; 0.444 for act, alt, and aerA respectively, while for ast and hlyA value was 0.957 in both cases (Table 1). On the other hand, differences were observed in the distribution of virulence genes and associated species p<0.01. The most prevalent gene was act in A. veronii and alt in A. caviae for p<0.01. All A. hydrophila studied were positive for the virulence genes ast and hlyA, while the aerA gene was only found in A. veronii and A. hydrophila although the difference found between these two species was not significant p=0.220 (Table 2).
Distribution of virulence genes in Aeromonas spp. strains.
| Total strains (n=218) | Virulence genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| act | alt | ast | hlyA | aerA | ||||||
| Co-pathogen | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) |
| Not (n=166) | 154 (76.2) | 12 (75.0) | 11 (68.8) | 155 (76.7) | 163 (76.2) | 3 (75.0) | 163 (76.2) | 3 (75.0) | 159 (75.7) | 7 (87.5) |
| Yes (n=52) | 48 (23.8) | 4 (25) | 5 (31.3) | 47 (23.3) | 51 (23.8) | 1 (25.0) | 51 (23.8) | 1 (25.0) | 51 (24.3) | 1 (12.5) |
| Total | 202 | 16 | 16 | 202 | 214 | 4 | 214 | 4 | 210 | 8 |
| Total strains (n=114) | Virulence genes in children | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| act | alt | ast | hlyA | aerA | ||||||
| Co-pathogen | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) | Not-presence (%) | Presence (%) |
| Not (n=70) | 43 (38.0) | 1 (100) | 3 (100) | 41 (36.9) | 44 (38.6) | 0 | 44 (38.6) | 0 | 44 (38.6) | 0 |
| Yes (n=44) | 70 (62.0) | 0 | 0 | 70 (63.1) | 70 (61.4) | 0 | 70 (61.4) | 0 | 70 (61.4) | 0 |
| Total | 113 | 1 | 3 | 111 | 114 | 0 | 114 | 0 | 114 | 0 |
Presence of virulence genes by species.
| Virulence genes | |||||
|---|---|---|---|---|---|
| Species (n=288) | act | alt | ast | hlyA | aerA |
| A. veronii (n=26) | 22 (84.6%) | 4 (15.4%) | 0 | 0 | 15 (57.7%) |
| A. hydrophila (n=16) | 6 (37.5%) | 16 (100%) | 16 (100%) | 16 (100%) | 6 (37.5%) |
| A. media (n=15) | 0 | 15 (100%) | 0 | 0 | 0 |
| A. salmonicida (n=1) | 1 (100%) | 1 (100%) | 0 | 1 (100%) | 0 |
| A. caviae (n=230) | 0 | 228 (99.1%) | 0 | 0 | 0 |
| Total | 29 | 262 | 16 | 17 | 21 |
Since we found no significant differences between strains isolated with a primary pathogen versus those isolated alone, we should think that they are all potentially just as virulent strains and that they must be treated with equal importance when treating the patient. But it is also true that the most prevalent species is A. caviae that, as we demonstrated, that is the one in which fewer virulence factors were detected, therefore, it seems potentially less aggressive. The study of expression or cytotoxicity of the positive alt gene in 99.1% of our isolates would be missing to complete this comparison to definitively verify whether there are differences between co-pathogenic isolates and single-isolation strains.
As for the proportion of genes present in each of the species is not in line with some published studies as Soltan Dallal et al., 201611 or Pablos et al., 201012 because, in our opinion, identifying isolates methods have improved regarding when the mentioned studies were done. Our study is according with results of Silva et al., 2017,13 who analyzed a greater number of strains and concluded that taking into account the number of virulence genes present in the majority of the isolates, the species A. hydrophila exhibit higher virulence potential among the species studied.
Not being able to clarify the controversy about the pathogenesis of Aeromonas, especially in pediatric patients, for not having been able to carry out a comparative study with strains isolated from healthy carriers, it would be prudent to analyze its appearance together with a compatible clinic. With all this, to say that antibiotic treatment would be indicated only in patients selected as some authors advised14 and, in this case yes, taking into account those species with a greater virulent potential as has been demonstrated.
In conclusion, A. hydrophila is the potentially most virulent species studied. The role of Aeromonas as a co-pathogen in feces, especially in children, should be further studied. And it would be useful to study the possible association of virulence expression with clinical symptoms in patients and also with healthy carriers.
Transparency declarationsAll authors have nothing to declare. This study has not been financially supported by any Diagnostic/Pharmaceutical company.
Ethical approvalNot applicable.
FundingThe authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Conflict of interestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
None.