Group A Streptococcus [GAS] is a community-acquired pathogen causing non-invasive and invasive infections, which are a major cause of morbidity and mortality worldwide.1 Healthcare-associated infections due to GAS and nosocomial transmission have been well described.2–4
Currently the most widely used method for GAS typing is 5′ emm gene sequencing which encodes the hypervariable region of the M protein, a major virulence determinant of GAS.5 At least 200 emm types have been defined (https://www.cdc.gov/streplab/groupa-strep/index.html) being emm1 and emm89 the most frequently associated with severe infections in Europe.6 Several streptococcal pyrogenic exotoxins (Spe) are implicated in the aggressiveness of GAS diseases. Spe genes (speA, speB, speC, speF, speG, speH,speJ, speK, ssa and smeZ) encode a group of mitogenic proteins secreted by many GAS strains, most of them showing very potent superantigen activity. Toxin-gene profiling is commonly used for GAS characterization.7
We report four invasive GAS infection cases in the Hospital Universitari Arnau de Vilanova (Lleida, Spain) in a short time: Patient A was a 65-year-old woman who required ICU admission on April 13, 2019 with septic shock; Patient B was a 67-year-old woman admitted in the ICU on April 26, 2019 with septic shock and pneumonia; Patient C was a 40-year-old immunosuppressed man who were admitted to the Emergency setting on April 28, 2019 with fever; Patient D was a 82-year-old woman admitted to the Emergency setting on May 28, 2019 with fever and surgical wound infection.
Three strains were isolated from blood samples (strains A, C and D) and one from bronchoalveolar lavage (BAL) (strain B).
GAS antibiotic susceptibility testing was performed with Panel Type 33 of Microscan WalkAway microdilution system using the breakpoints recommended by The European Committee on Antimicrobial Susceptibility Testing.8
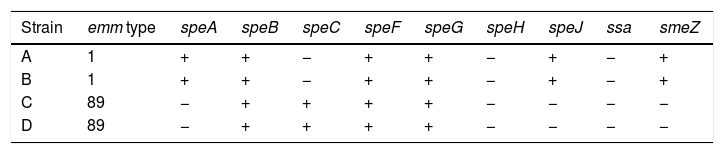
Because the four invasive GAS cases [three bacteremias (patient A, patient C and patient D) and one pneumonia (patient B)] occurred so closely in time, the isolates were sent to the Centro Nacional de Microbiología, Instituto de Salud Carlos III for M protein gene (emm) typing and toxin-gene profiling as previously described.7
The four GAS strains were susceptible to penicillin, clindamycin, erythromycin, tetracycline and vancomycine. Toxin-gene profiles and emm typing of the four GAS strains are shown in Table 1.
All strains harbored the 3 superantigenic toxin genes: speB, speF and speG regardless of emm type. GAS strains with the same emm type have shown the same toxin profile (Table 1) coinciding with that described by Bencardino D et al.9
An identical clone emm1 was isolated from two of the patients (A and B) coinciding in time in ICU setting (April 26–29, 2019). Strain A was recovered from blood sample obtained in the Emergency Service previously ICU admission. Strain B was isolated from a BAL obtained upon ICU admission. Both patients developed a serious invasive infection with multiorgan failure and were treated with penicillin and clindamycin after GAS isolation. Patient A was discharged from ICU setting after 16 days of admission (April 29, 2019) and patient B died after 4 days of admission (April 30, 2019). A healthcare associated GAS infection has been discarded due to BAL sample of patient B was obtained in the first day of ICU admission.
Strains from patients C and D were an identical clone emm89. The emergence of emm89 strains has been described in several geographic locations.6 Blood samples of both patients were obtained in the Emergency setting upon admission and patients did not coincide in time in our hospital. Both patients were treated with penicillin and clindamycin after GAS isolation and the evolution of both patients was favorable.
ConclusionsIn our study the GAS clone emm1 has been related with severe invasive infections and was isolated from two patients who were coinciding in time in ICU setting but nosocomial transmission was discarded, while the GAS clone emm89 was isolated from two patients with a lighter invasive GAS infection that did not coincide in time in hospital. More molecular studies are needed to describe the molecular epidemiology of GAS infection in our area and the most prevalent GAS circulating clones.
Conflict of interestThe authors declare no conflict of interest.