Triple negative breast cancer (TNBC) is a subtype of breast cancer (BC) with a heterogeneous nature that stains negatively for estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor 2 (HER2) during immunohistochemistry. Approximately 15–20% of all cases of breast cancer are triple negative phenotypes. Compared to patients with hormone receptor-positive cancer, TNBC patients are typically younger (<50 years), African American, and have a high incidence of mutations in BRCA1/2 genes. To date, not a single targeted therapy has been approved for TNBC treatment, and cytotoxic chemotherapy remains as the standard systemic treatment, meaning that TNBC is an aggressive subtype of breast cancer with a poor prognosis. In this review, the literature search was done up to date on which gene expression profile of TNBC has been analyzed in order to identify the consensus on molecular mechanisms involved in carcinogenesis and/or the prognostic markers of the disease. In conclusion, these studies have reported that TNBC is composed of several clusters or genomic signatures as basal keratins. They have also reported on their proliferation, luminal/basal apocrine, regulatory interferon, immune cells/immunoglobulin related to stem cells, epithelial-mesenchymal, androgen receptor and angiogenesis. However, not all research groups have reported reproducible results. This confirms the heterogeneous nature of TNBC and the need for research on uniform selection criteria. However, these discoveries have led to the proposal of new treatments, such as the addition of platinum salts, new combinations of therapeutic agents, some targeted therapies such as PARP inhibitors, and PI3K and androgen antagonists. There is no doubt that a better understanding of the nature of TNBC will allow individualized and more effective therapies.
Breast cancer (BC) is the main cause of death by cancer among women, and represents 30% of all new cancer cases in the Caucasian population. A woman living in the United States has a 12.3% (1 in every 8) risk of being diagnosed with breast cancer.1 According to the World Health Organization, 1.67 million new cases were registered worldwide in 2012.2 In Mexico, BC is also the first place in malignant neoplasia incidence among women. It represents 11.34% of all cancer cases, with a global increase of approximately 1.5% annually. However, in emerging countries this increase is up to 5%.3
Despite having the same origin tissue, BC represents a heterogeneous cancer group with complex biological behavior and a great clinical variability. Over the last 10 years, extensive research at a molecular and genetic level has been conducted in order to sub-type these BCs. This has permitted the determination of clinical, pathological and molecular variables, to select treatment modalities and forecast, in some cases, the evolution of the disease at the moment of diagnosis.4
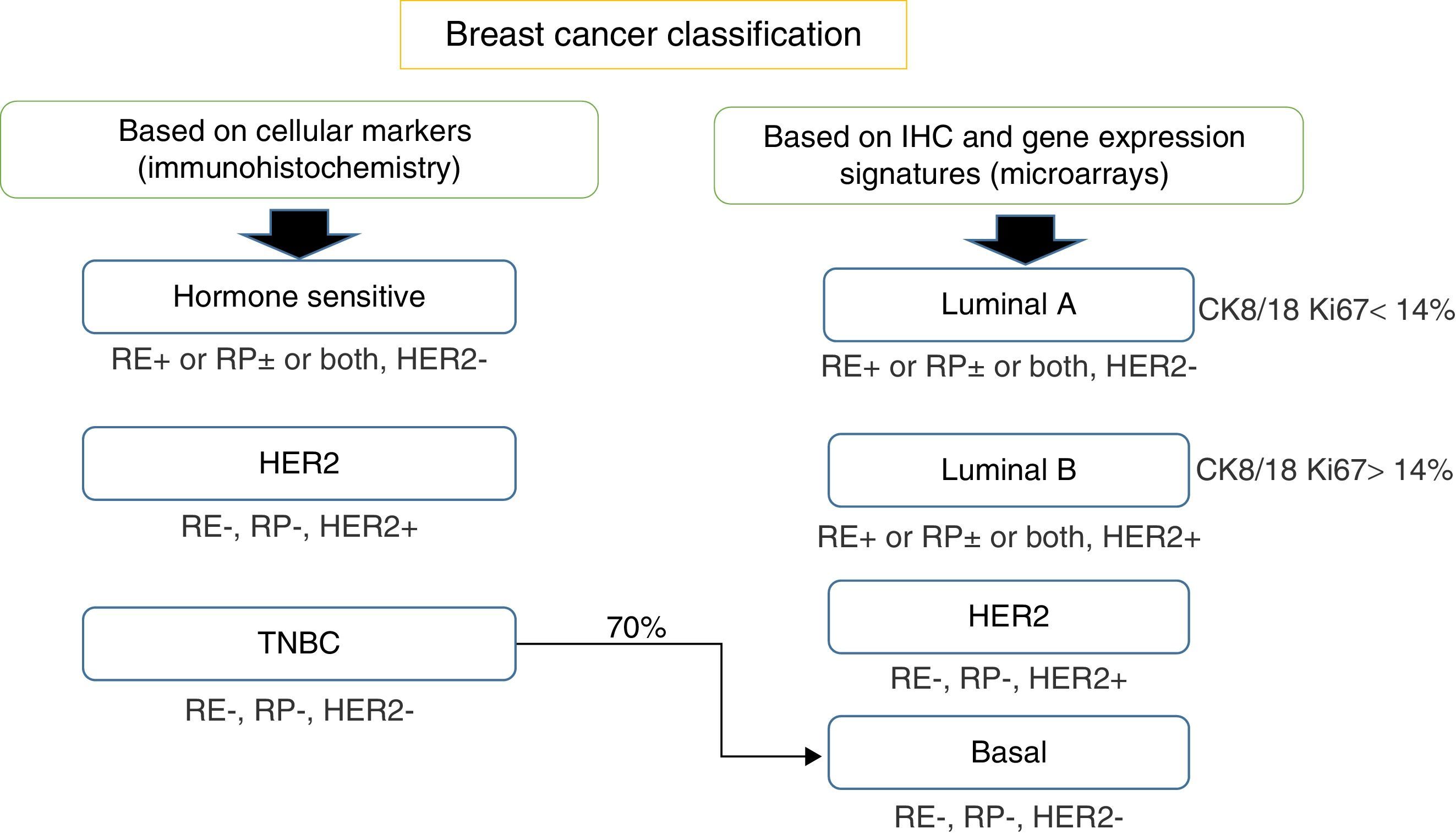
Breast cancer classificationIn the traditional way the most important information that pathologists gave oncologists, respect to the classification of breast cancer, included the status of the nodules and tumor size, histological grade and standard immunohistochemistry tests (IHC) status of hormone receptors: estrogen receptor (ER) and progesterone receptor (PR) (Fig. 1).
Breast cancer classification according to immunohistochemistry cellular markers (IHC) or according to a combination of IHC and microarray expression methods (gene signatures). ER, estrogen receptors; PR, progesterone receptors; HER2, epidermal growth receptor 2; CK, cytokeratin; EGFR, growth factor receptor.
Later on, in the era of trastuzumab, information on the amplification status of the human epidermal growth factor receptor 2 gene (HER2) became routine. Combining all these parameters, it was possible to classify patients as high or low risk. There were however a group of patients who were in an intermediate category that could not be classified between these two groups. Results showed that 15% of the patients classified as low risk had relapsed or died due to a very aggressive disease. And patients classified as high risk, surprisingly 10–15% had a favorable response. All those results led to the conclusion that the method of classification was not very appropriate.
Thus, based only on standard IHC directed at cellular markers that reflect the availability of targeted therapies, breast cancer can also be classified into three main groups: (a) homone sensitive (ER or PR positive), (b) HER2 positive, sensitive to trastuzumab or (c) triple negative breast cancer (TNBC), defined by the absence of ER, PR and HER2 amplification.5–8
No targeted treatment is available for TNBT and chemotherapy remains the best therapeutic option. However, in the case of recurrence or chemoresistance, therapeutic options are very limited.9
Subsequently, in the late 90s, when platforms for genomic studies were available, IHC methods coupled with complementary DNA (cDNA) microarray technology, allowed for a more extensive BC classification and were able to define four different BC subgroups, which differ in prognosis and targets: (a) luminal A – positive to ER and PR, negative to the amplification of HER2, with a low proliferation index (Ki-67<14%); (b) luminal B – ER positive, HER2 positive or negative, high proliferation index (Ki-67>14%), negative or low positive PR; (c) positive for HER2 – overexpressed or amplified HER2, negative ER and PR; (d) basal-like or TNBC, also defined by the absence of ER, PR and HER2 amplification. However, this last assumption is not strictly accurate due as not all basal-like tumors are completely TNBC.10,11
This classification is based on the consideration that there are two cell types in the mammary gland, luminal cells and basal cells. These two cell types can be differentiated by IHC tests as luminal cells, which express ER and PR and that are positive for keratins 8/18, while basal cells are positive for keratins 5/6 and 17.12,13
Because of the cost involved in utilizing microarray technology in the clinic, the most employed technique for BC classification remains IHC and FISH.6,8,14
Between 60% and 70% of all breast cancers correspond to positive ER and PR subtypes,6 which usually respond to targeted ER therapy, such as selective ER modulators (i.e. tamoxifen, or aromatase inhibitors) which reduce serum estrogen concentrations. Around 15–20% of BCs amplify or overexpress the HER2 oncogene. This type of cancer is associated with an incorrect prognosis and the treatments specifically aimed at HER2 (i.e. trastuzumab) have greatly improved results in women with BC HER2+.15
Prognostic value of genomic signatures in BCDespite the fact that they have not completely turned into routine methods, genomic methods have been used in research studies with the purpose of: (a) getting a better understanding of the nature of these variations of BC; (b) stratifying patients through assessment of the evolution of the disease (evaluating clinical pathological characteristics), and (c) understanding the possible response of the patient to treatment (in relationship with ER, PR, HER2 and the proliferation rate).5,16
Through global expression analysis with microarrays, the identification of gene groups (genomic signatures) has been possible, which allow us to estimate risk of recurrence of the disease and/or predict the response to adjuvant therapy in BC patients in the early stages.17–20 Some of these genomic signatures have been validated in large cohorts and have been the precedent for some commercial genomic tests, providing complementary information to clinicians in order to obtain a more precisely classification patients who has a high risk of recurrence, and to offer more personalized attention.
The first commercial signature, and the one still widely distributed under its commercial name, is MammaPrint® (Agilent, Amsterdam, Holland). This signature measures mRNA of 70 gene expression and was approved by the US FDA and by regulators in the EU as an essay with prognostic value in patients with BC who are under 61 years of age, in an I/II stage, with negative lymph nodes or one to three positive lymph nodes. In addition to accurately identifying patients who can safely avoid adjuvant chemotherapy.17,21 MammaPrint stratifies patients into low-risk or high-risk groups.17 However, even though discrimination is good for ER+ cancers, it is not the same with ER− cancers,22,23 limiting its clinical value to a subgroup of patients.
Another widely distributed and commercially available assay is Oncotype DX® test (Genomic Health, Redwood City, CA, U.S.). This test is used to classify patients with Luminal A and B BCs, as well as HER2+ subtypes (though not TNBC), since it has been designed to assess genes related to ER, proliferation genes, HER2 genes, and genes related to invasion, among others.19 This test allows the oncologist to discern whether or not chemotherapy treatment will be beneficial. This essay measures the 21-gene expression, which provides information of the recurrence of the disease through a scoring system from 0 to 100, stratifying patients into low-risk (score <18), intermediate risk (score 18–30) or high risk (≥31) groups.19,20
A third test, consisting of an algorithm for the intrinsic molecular classification of BC, has been nominated PAM50 (Fig. 2). This was designed to improve IHC and microarray classification aggreance.24 This 50-gene signature can classify BCs as luminal A, luminal B, HER2 and basal-like. The PAM50 score was designed with the purpose of translating the different intrinsic subtypes into an associated prognostic value.24 An application of this score is the identification of patients who may benefit from the weekly addition of paclitaxel to conventional chemotherapy with anthracycline as an adjuvant treatment of operable BC with positive lymph nodes.25 In 2013, Prosigna (Seattle, Washington, U.S.) started commercializing a diagnostic kit which qualifies mRNA expression of the 50 genes used by the algorithm in order to calculate the risk of recurrence.26
In addition to these tests, which are the most commonly distributed, there are others in the market,27–29 where the common denominator is the measurement of gene expression of proliferation related genes. It is important to mention that these multiple-gene assays are for the most part, if not exclusively, applicable to luminal BC, thus stressing once more the need to search for and identify markers with prognostic value and response to treatment in BC patients with negative hormonal receptors or HER2 amplification.
Triple negative breast cancer (TNBC)Since 1997, the National Comprehensive Cancer Network (NCCN) has gathered data of women with newly diagnosed breast cancer, representing many institutions around the U.S. HER2 by IHC's status was added to the data from the NCCN as a routine element in the year 1999, and HER2 status through fluorescent in situ hybridization (FISH) was added in 2001. The term “Triple Negative Breast Cancer” (TNBC) appeared for the first time in the literature in 2005.30 Due to the lack of effective therapeutic options, many studies have focused on the understanding of the biology of TNBC for the selection of biomarkers and therapeutic targets. In this review, we will focus on making a compilation of molecular and gene studies in reference to this BC subgroup.
TNBC risk factorsTNBC makes up about 15–20% of all BCs and follows an aggressive clinical course, including a high incidence of visceral and nervous metastases, compared to BCs positive to hormone receptors, TNBC is more frequently associated with BRCA1 and, to a lesser extent, BRCA2 mutations.31,32 It has also been associated with a high local recurrence rate at three years after treatment, and a high death rate at 5 years. Upon long-range metastasis, the survival rate is poor.33 Differently from other subtypes of BC (Luminal A, B and HER2), where early pregnancy has been recognized as a protective factor against BC, gestation appears to be an important risk factor in the development of the triple negative phenotype. In a study by Phipps et al.,34 they reported that women who had more children (three or more) had a greater risk (1.4 times) of TNBC. Later, a study by Tamimi et al.35 reaffirmed that multiple pregnancies were associated with an increased risk of developing TNBC.
Other behaviors associated with the triple negative subtype are a lack of lactation,36 and various studies have made an association between TNBC and a high body mass index (BMI), a metabolic syndrome, type 2 diabetes, obesity and/or insulin resistance.37–43 Consequently, patients with TNBC are the group with the worst prognosis, which is important to consider in a country like Mexico, which has an elevated prevalence of obesity and metabolic syndrome.
Racial ancestry has been shown to have an important relation to TNBC, since 21% of Afro American BCs have been classified as TNBC (compared to 15% of the Caucasian population).44–46 The Hispanic population has also shown a high incidence of this cancer subtype, with a frequency of 21.3% in Peru,47 27% in Brasil,48 24.1% in Venezuela49 and 23.1% in Mexico.50
TNBC presents a high rate of proliferation51 and is highly aggressive.32,52–54 In a study by Sorlie et al., in which the response to an uniform treatment was evaluated, they reported differing results between patients belonging to groups of distinct subtypes. Among these results exist a report that patients with TNBC had the worst prognosis by a significant margin when compared to patients with cancers that had positive hormone receptors.11 In a later study, Dent et al. compared the clinical history, outcome and the nature of the disease in women with TNBC to women with other subtypes. The results showed that TNBC had a low survival rate and a high rate of long-distance metastasis after treatment.52 Due to TNBC's aggressive course, these types of cancers can be highly sensitive to cytotoxic medications. The proportion of patients who reach a complete pathological response (CPR) after treatment were found to be 30–45%,55,56 and the patients, which achieved a CPR had excellent long-term results, with a global survival rate higher than 90%. In addition, if CPR is achieved, patients with TNBC or other types of BC had a similar survival rate. However, those patients with TNBC who presented with a residual disease had worse results than patients with other types of BC.45,57 The aggressiveness of TNBC and the shortage of specific therapeutic options underline the need to understand the ways in which this type of cancer develops.
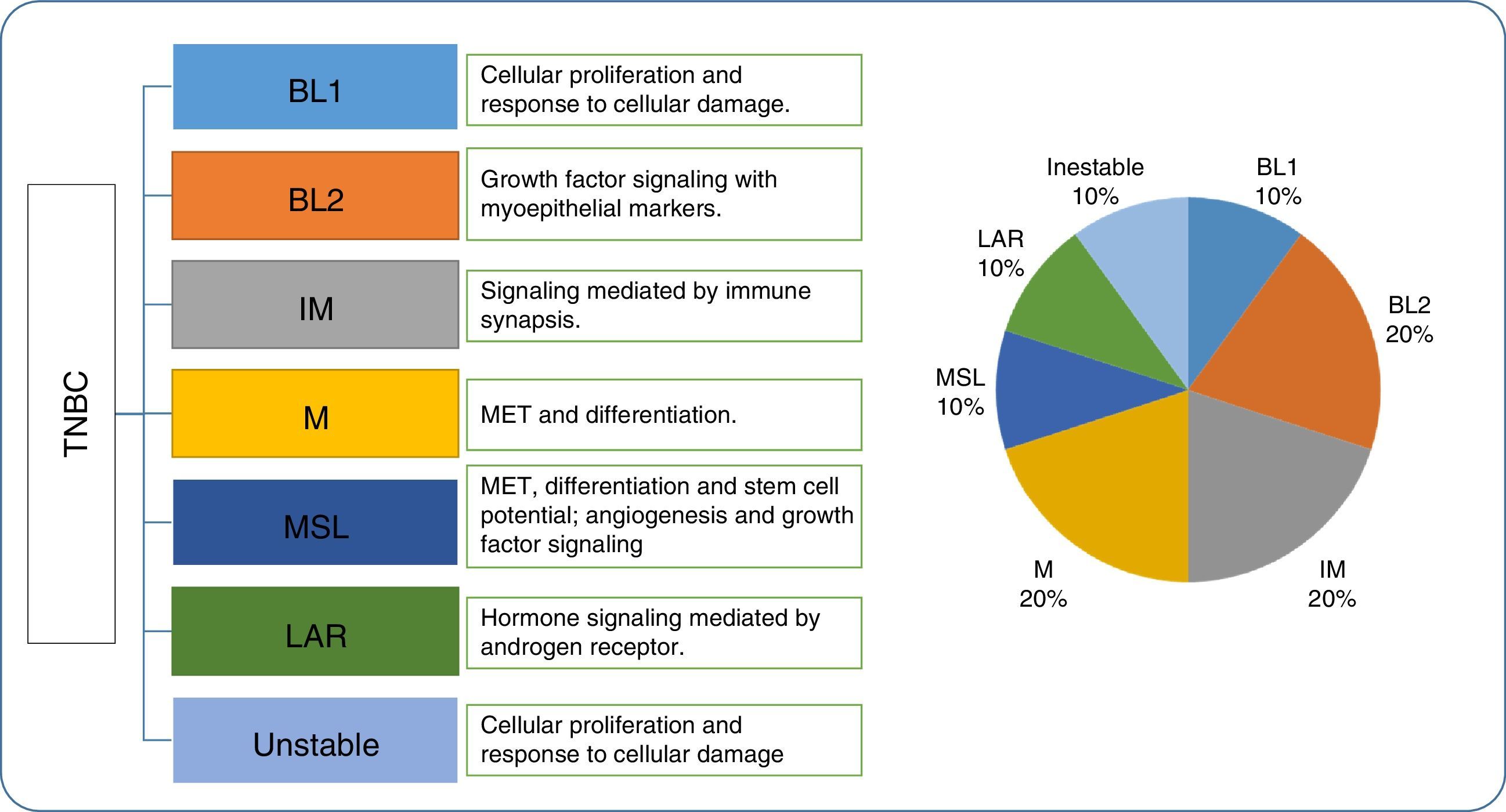
Molecular classification of TNBC subtypesIn 2007, Kreike et al. reported that TNBCs were synonymous with basal type molecular cancers.58 However, as we mentioned before, it has been proven that TNBC's molecular biology is diverse and heterogenous,59 and that TNBC is not an ideal substitute for the basal phenotype. In 2011, Lehmann et al.60 identified 6 subtypes of TNBC, as well as 1 unstable subtype. The six subtypes included two basal like (BL1 and BL2), one immunomodulatory class, one mesenchymal class, one mesenchymal stem cell (MSC) class, and one luminal androgen receptor (LAR) class. In addition, gene expression analysis allowed the authors to identify the cell lines representative of each TNBC subtype to later evaluate their response to different medications.
BLI: Cancers in this group are characterized by their rapid cell division, with an increased proliferation rate and loss of cell cycle control. Due to this, they present an elevated expression (mRNA) of Ki67 and respond to antimitotic agents.
BL2: These cancers present the expression of EGFR, TP63, MET, etc. and involve activation of glycolysis and gluconeogenesis routes.
IM: They are composed of response cells from the immune system, and present antigens and cytokine expression. They overlap with medullary breast cancer with a favorable prognosis.
M and MSL: These groups present epithelial-mesenchymal transition markers, including angiogenesis (VEGFR2) and most likely respond to desatinib (TK inhibitors) and mTOR.
LAR: This group is characterized by the expression of androgen receptors (AR). They are RE negative, but maintain an elevated hormonal component, regulated by steroid synthesis, porphyrin metabolism genes and androgens/estrogens. The AR's expression is up to nine times more than the other groups, and are prime candidates for anti-androgen therapies.
The signaling pathways obtained by the study are potential pharmacological targets in these cell models to that analysis of these distinct genetic signatures can be used to select a therapy. In general, it was observed that the BL1 and BL2 subtypes respond well to cisplatin, and that the M and MSL types respond well to NVP-BEZ235, a PI3K/mTOR inhibitor and dasatinib (a tyrosine kinase inhibitor). The LAR subtype was only sensitive to bicalutamide (an androgen receptor agonist).
Later, Burstein et al.61 confirmed four of the subtypes reported by Lehmann with the base DNA profiles or the single nucleotide DNA markers from 198 TNBC samples: one luminal androgen receptor type, one mesenchymal type, and two basal types. These studies by Burstein and Lehmann suggested the existence of at least 4 TNBC subtypes (Fig. 3).
Lehman's triple negative breast cancer classification, gene ontology and proportion. TNBC, triple negative breast cancer; BL1, basal like 1; BL2, basal like 2; IM, immunomodulatory; M, mesenchymal; MSL, mesenchymal stem cell-type; LAR, androgen luminal receptor; MET, mesenchymal epithelial transition.
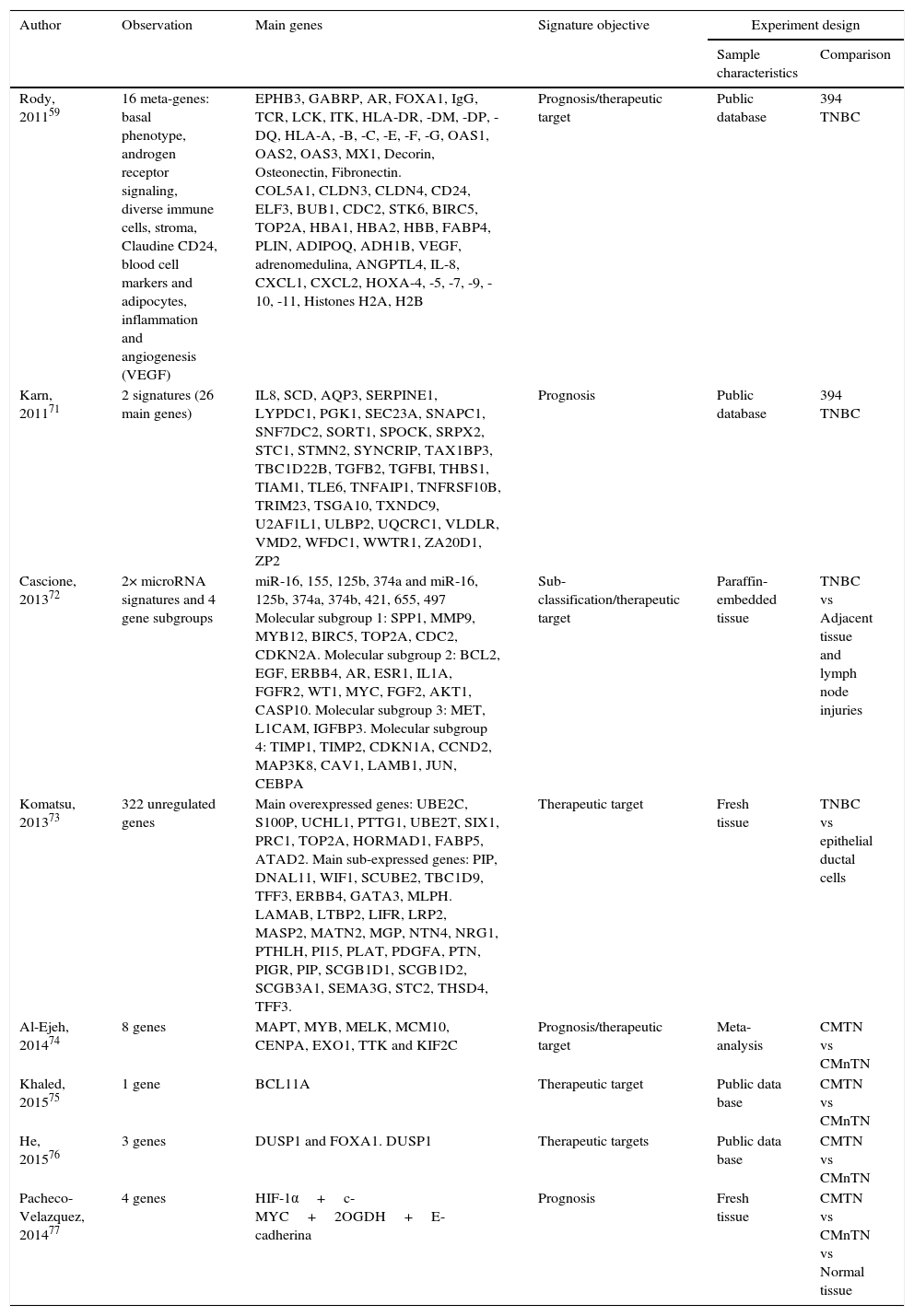
Various expression studies have been performed on TNBC with the aim of identifying therapeutic targets, which have been summarized in Table 1. This summary includes the characteristics of each study and the selected differentially expressed principal genes. This table shows the variability of the data obtained from each study. This indicates that validation of these possible molecular targets is necessary and even more studies are required to find efficient biomarkers for the treatment of TNBC.
Main genes reported from researches which analyze TNBC gene expression profiles. TNBC, triple negative breast cancer; nTNBC, non-triple negative breast cancer.
| Author | Observation | Main genes | Signature objective | Experiment design | |
|---|---|---|---|---|---|
| Sample characteristics | Comparison | ||||
| Rody, 201159 | 16 meta-genes: basal phenotype, androgen receptor signaling, diverse immune cells, stroma, Claudine CD24, blood cell markers and adipocytes, inflammation and angiogenesis (VEGF) | EPHB3, GABRP, AR, FOXA1, IgG, TCR, LCK, ITK, HLA-DR, -DM, -DP, -DQ, HLA-A, -B, -C, -E, -F, -G, OAS1, OAS2, OAS3, MX1, Decorin, Osteonectin, Fibronectin. COL5A1, CLDN3, CLDN4, CD24, ELF3, BUB1, CDC2, STK6, BIRC5, TOP2A, HBA1, HBA2, HBB, FABP4, PLIN, ADIPOQ, ADH1B, VEGF, adrenomedulina, ANGPTL4, IL-8, CXCL1, CXCL2, HOXA-4, -5, -7, -9, -10, -11, Histones H2A, H2B | Prognosis/therapeutic target | Public database | 394 TNBC |
| Karn, 201171 | 2 signatures (26 main genes) | IL8, SCD, AQP3, SERPINE1, LYPDC1, PGK1, SEC23A, SNAPC1, SNF7DC2, SORT1, SPOCK, SRPX2, STC1, STMN2, SYNCRIP, TAX1BP3, TBC1D22B, TGFB2, TGFBI, THBS1, TIAM1, TLE6, TNFAIP1, TNFRSF10B, TRIM23, TSGA10, TXNDC9, U2AF1L1, ULBP2, UQCRC1, VLDLR, VMD2, WFDC1, WWTR1, ZA20D1, ZP2 | Prognosis | Public database | 394 TNBC |
| Cascione, 201372 | 2× microRNA signatures and 4 gene subgroups | miR-16, 155, 125b, 374a and miR-16, 125b, 374a, 374b, 421, 655, 497 Molecular subgroup 1: SPP1, MMP9, MYB12, BIRC5, TOP2A, CDC2, CDKN2A. Molecular subgroup 2: BCL2, EGF, ERBB4, AR, ESR1, IL1A, FGFR2, WT1, MYC, FGF2, AKT1, CASP10. Molecular subgroup 3: MET, L1CAM, IGFBP3. Molecular subgroup 4: TIMP1, TIMP2, CDKN1A, CCND2, MAP3K8, CAV1, LAMB1, JUN, CEBPA | Sub-classification/therapeutic target | Paraffin-embedded tissue | TNBC vs Adjacent tissue and lymph node injuries |
| Komatsu, 201373 | 322 unregulated genes | Main overexpressed genes: UBE2C, S100P, UCHL1, PTTG1, UBE2T, SIX1, PRC1, TOP2A, HORMAD1, FABP5, ATAD2. Main sub-expressed genes: PIP, DNAL11, WIF1, SCUBE2, TBC1D9, TFF3, ERBB4, GATA3, MLPH. LAMAB, LTBP2, LIFR, LRP2, MASP2, MATN2, MGP, NTN4, NRG1, PTHLH, PI15, PLAT, PDGFA, PTN, PIGR, PIP, SCGB1D1, SCGB1D2, SCGB3A1, SEMA3G, STC2, THSD4, TFF3. | Therapeutic target | Fresh tissue | TNBC vs epithelial ductal cells |
| Al-Ejeh, 201474 | 8 genes | MAPT, MYB, MELK, MCM10, CENPA, EXO1, TTK and KIF2C | Prognosis/therapeutic target | Meta-analysis | CMTN vs CMnTN |
| Khaled, 201575 | 1 gene | BCL11A | Therapeutic target | Public data base | CMTN vs CMnTN |
| He, 201576 | 3 genes | DUSP1 and FOXA1. DUSP1 | Therapeutic targets | Public data base | CMTN vs CMnTN |
| Pacheco-Velazquez, 201477 | 4 genes | HIF-1α+c-MYC+2OGDH+E-cadherina | Prognosis | Fresh tissue | CMTN vs CMnTN vs Normal tissue |
TNBC is a subtype of BC with a very heterogeneous behavior which, based on gene expression analysis by microarray, was classified by Lehmann into six distinct groups. This sub-classification is not only useful to the better understanding of this disease, but also to identify new molecular targets for its treatment. Following are some of the proteins which have been identified in these TNBC groups as potential therapeutic targets.
EGFR: This is one of the principal receptors, and plays an important role in the survival of the tumor. It is a marker of cellular proliferation, angiogenesis, metastasis and apoptosis inhibition.
VEGF: This is the principal marker for angiogenesis. It induces proliferation and maintains the integrity of the cell.
C-kit and basal cytokeratins: C-kit unites with the receptor of a growth factor, stimulates survival and differentiation, and induces invasiveness in the cancerous cell. The basal cytokeratins are intermediate filament proteins. Their expression is constant when epithelium undergoes transformation.
P53: suppressor protein important in cell cycle regulation and tumor apoptosis.
hKi67: A cellular proliferation marker which is present in the nucleus during interphase and mitosis. Ki67 expression in normal mammary tissue is <3%. The expression of TNBC can elevate it up to 80%. It self-expresses particularly in RE-negative cells. It is associated with an unfavorable prognosis.
PARP: A family of cell signaling enzymes. 18 PARP proteins have been identified. PARP1 is responsible for the majority of its functions. It is a DNA damage sensor and plays an important role in its repair and in responses to inflammation, ischemia and oxidative stress. In cells with a BRCA mutation, PARP inhibition contributes to cell death by apoptosis.
Hsp90: A cellular chaperone, it stabilizes the conformation of many labile proteins, such as steroid receptors, CDK4, RAF-1 and AKT, etc. PU-H71 is a cyclin inhibitor which has been shown to be useful in some TNBC models.
COX2: This protein is expressed by the inflammatory stimulus and its expression is associated with an unfavorable prognosis.
TK: A regulatory protein which helps the cell grow and differentiate itself.
mTOR: The over-expression of these proteins (PI3K/mTOR) is associated with a poor response to treatment with hormones and trastuzumab.
Women with TNBC are not able to take endocrinology therapies neither HER2 targeted therapies, hence treatment options are limited to surgery, radiotherapy and chemotherapy,62,63 being chemotherapy the standard treatment of TNBC. In the past two decades the use of more aggressive therapy has produced a clearer improvement in quality of life; however, there is a consensus opinion, which considers that it would be impossible to improve these results if this cytotoxic therapy remained the only option to be offered to these patients. For this reason, there is an urge to find new “intelligent drugs” capable of solving chemoresistance and reducing the risks of chemotherapy in patients who do respond. The greatest obstacle to find actionable targets is the vast heterogeneity both inter and intra-tumor of the TNBC. In fact, several years of study have failed to prove a unified alteration which may serve as targets of a targeted therapy.
Several researches about TNBC describe genomic signatures involving affected signaling pathways, including cellular, immunologic and metabolic processes like – basal keratin differential expressions, interferon proliferation and regulation routes, low expression in Claudine, mesenchymal epithelial transition, androgen receptor expression (AR) and angiogenesis activation (VEGF).58–60 These findings have revealed potential molecular targets,49 and different drug types are currently found under investigation, which can basically be defined in four groups: (1) agents which cause damage to DNA (i.e. cisplatin, cyclophosphamide), (2) agents which inhibit poly (ADP-ribose) polymerase (PARP inhibitors), (3) tyrosine-kinase inhibitors and (4) agents which inhibit downstream signaling pathways (mainly PI3K/AKT).64 Down below there are the descriptions of treatments which have been proven depending upon identified therapeutic targets in TNBC.
- •
Bevacizumab: Anti-VEGF (vascular endothelial growth factor) monoclonal antibody that blocks angiogenesis; although, a lack of efficiency as well as a toxicity increment have been observed.65
- •
Olaparib/iniparib: PARP inhibitors in patients with mutations in BRCA1/2, they have also been widely studied since they have shown to have anticancer selective activity in BRCA1 and BRCA2-deficient cancers; nevertheless, the relevance of the inhibition of these enzymes has yet to be confirmed.66
- •
Cetuximab: Anti-EGFR monoclonal targeted antibody, it has been evaluated as monotherapy or with a platinum-based therapy (carboplatin). Even though, at first, this therapy seemed to be very promising, it has proven very modest results.67
- •
mTOR inhibitors (mammalian target of rapamycin) – mTOR is a PI3K signaling pathway effector regulated by AKT and suppressor of PTEN tumors. PI3K pathway proteins are frequently affected by mutations in mammary carcinomas, and the loss of PTEN is a frequent discovery in TNBC; thus, increasing the activation of mTOR in the diseases.68 It is for this reason that today, mTOR inhibitor drugs have been under evaluation.69
- •
Bicalutamide/enzalutamide (“anti-androgen” targeted therapy): Some preclinical studies showed that luminal TNBC which expresses the androgen receptor (AR) is sensitive to androgen deprivation. Bicalutamide as monotherapy demonstrated a clinical benefit of 19% of the AR positive.70 Today, a phase II study is evaluating enzalutamide's safety and efficacy in TNBC and AR positive patients. Results are yet to be seen.
Targeted therapies against some identified biomarkers in TNBC have not proven a significant improvement, the problem has been the lack of dependable predictive biomarkers, which is essential before any of these treatments can be introduced in clinical practice.
ConclusionsTNBC is usually the most aggressive type of cancer and it is linked to the highest mortality rate in comparison to other types of BC. Numerous advances in the understanding of this cancer have been made. Reported genomic signatures to date reflect the heterogeneity of the triple negative phenotype, at the same time have allowed for a TNBC classification into 6 clinically relevant subgroups. One of these subgroups has a strongly immune component. Another subgroup shares biological characteristics with mutations in genes BRCA1/2 with deficiency in DNA repair.
While several TNBC biomarkers are reported in the literature every year, only few have exceeded validation to justify its use in clinical practice. The reason for this deterioration may be due to the fact that while strong associations often derive initially between the expression of a putative biomarker and the stage of the disease, candidate markers do not often have any relevant and significant functions of the disease in clinical laboratory and therapeutic practice. For TNBC specifically, different research groups that used the information generated by microarrays obtained results varying from one group to another and in some cases these results have not been able to be validated. This could be a result of the inherent heterogeneity of TNBC or each experiment design's own limitations, such as: (1) the use of large data base retrospective cohorts, where TNBC's classification is based solely on the information obtained from the gene expression profile data, this methodology is not the standard in clinical practice. Also, this information shows limitations due to how varying the data collection and sample management methods are used (in some cases paraffin-embedded tissue was utilized, others utilized fresh tissue, etc.) to feed the databases that are afterwards referenced by other researchers; (2) limited gene expression analysis of a determined number of genes, instead of conducting a global analysis of gene expression; (3) previous works and researches compared TNBC gene expression profiles vs other BC subtypes, or TNBC vs normal tissue, which does not unify stratification criteria.
In Mexico, there are few research papers which report the analysis of TNBC expression profiles This is concerning, since this type of BC jeopardizes a considerable amount of cases among the Mexican population, hence it is necessary to increase efforts in the search of biomarkers or possible therapeutic targets which can be validated in our population.
FundingCONACyT Fondo Salud162301 and Convocatoria de Becas Nacionales2014-370024.
Conflict of interestThe authors have no conflicts of interest to declare.