Epigenetics is defined as the study of the mechanisms that regulate gene expression without altering the underlying DNA sequence. The best known is DNA methylation. Multiple sclerosis (MS) is a disease with no entirely known aetiology, in which it is stated that the involvement of environmental factors on people with a genetic predisposition, may be key to the development of the disease. It is at this intersection between genetic predisposition and environmental factors where DNA methylation may play a pathogenic role.
DevelopmentA literature review of the effects of environmental risk factors for the development of MS can have on the different epigenetic mechanisms as well as the implication that such changes have on the development of the disease.
ConclusionKnowledge of epigenetic modifications involved in the pathogenesis of MS, opens a new avenue of research for identification of potential biomarkers, as well as finding new therapeutic targets.
La epigenética se define como el estudio de los mecanismos que regulan la expresión génica sin modificar la secuencia de ADN, siendo entre ellos el más conocido la metilación del ADN. La esclerosis múltiple (EM) es una enfermedad de etiología no del todo conocida, en la que se plantea que la participación de factores ambientales sobre individuos con una determinada predisposición genética, pueden resultar claves para el desarrollo de la enfermedad. Es en esta intersección entre la predisposición genética y los factores ambientales donde la metilación del ADN puede desempeñar un papel patogénico.
DesarrolloRealizamos una revisión bibliográfica de los efectos que los factores de riesgo ambiental para el desarrollo de EM pueden ejercer sobre los distintos mecanismos epigenéticos, así como la implicación que presentan dichas modificaciones en el desarrollo de la enfermedad.
ConclusiónEl conocimiento de las modificaciones epigenéticas involucradas en la patogenia de la EM abre una nueva vía de investigación para la identificación de potenciales biomarcadores, así como para la búsqueda de nuevas dianas terapéuticas.
The term “epigenetics” appeared in the literature for the first time in the mid-20th century (Conrad Waddington, 1905-1975),1 but it was not until recently that it has become an emerging research field. It is now considered a promising source of knowledge, especially in the medical field.
Epigenetics is the study of mechanisms regulating gene expression without modifying the deoxyribonucleic acid (DNA) sequence. This discipline represents a link between genetic and environmental influences on phenotype development. Epigenetic changes modify the activation of some genes depending on external conditions, and they are essential in cellular and tissue differentiation, which takes place during foetal development. These changes also occur during adulthood. Human cells experience epigenetic changes during their lifetimes. In fact, identical twins (with the same genetic load) accumulate different epigenetic patterns depending on the environmental factors to which they are exposed, for example, tobacco, diet, or exercise.2 This translates into observable differences in the phenotypes of both twins, which manifest as different susceptibilities to disease or disease outcomes.3
The main epigenetic mechanisms involve DNA methylation, histone modification, and the action of non-coding RNA. DNA methylation is the best-known mechanism and its association with the development of diseases has been the subject of many studies. This mechanism is the focus of our study.
DNA methylationDNA methylation is a process by which methyl groups are added to cytosine residues in the RNA nucleotide chain. This binding occurs at cytosine-guanine dinucleotides (CpG), which accumulate in the genome to form CpG islands. These islands are especially abundant in gene promoter regions and other regulatory regions. Methylation is carried out by DNA methyltransferases (DNMT) that catalyse the transfer of a methyl group from S-adenosyl-l-methionine (SAM) to carbon 5 of cytosine.4 This process may follow one of 2 models: establishing a de novo DNA methylation pattern catalysed by DNMT3a and DNMT3b,5 or maintaining a genomic methylation pattern through successive DNA replication cycles mediated by DNMT1. Methylation occurs during DNA replication such that when a CpG sequence adopts a certain methylation pattern, this modification becomes stable, and therefore inherited during DNA replication and maintained in daughter strands.6
Hypermethylation at CpG islands in gene promoter regions is typically a gene repression mechanism since it inhibits transcription. This inhibition takes place through 2 processes: a direct one which prevents binding of transcription factors containing recognition sites for methylated CpG, and an indirect one, which blocks the access to regulatory elements, necessary for transcription factor binding, by adhering protein complexes known as methyl-binding domains (MBD), which bind to methylated CpG sites.7
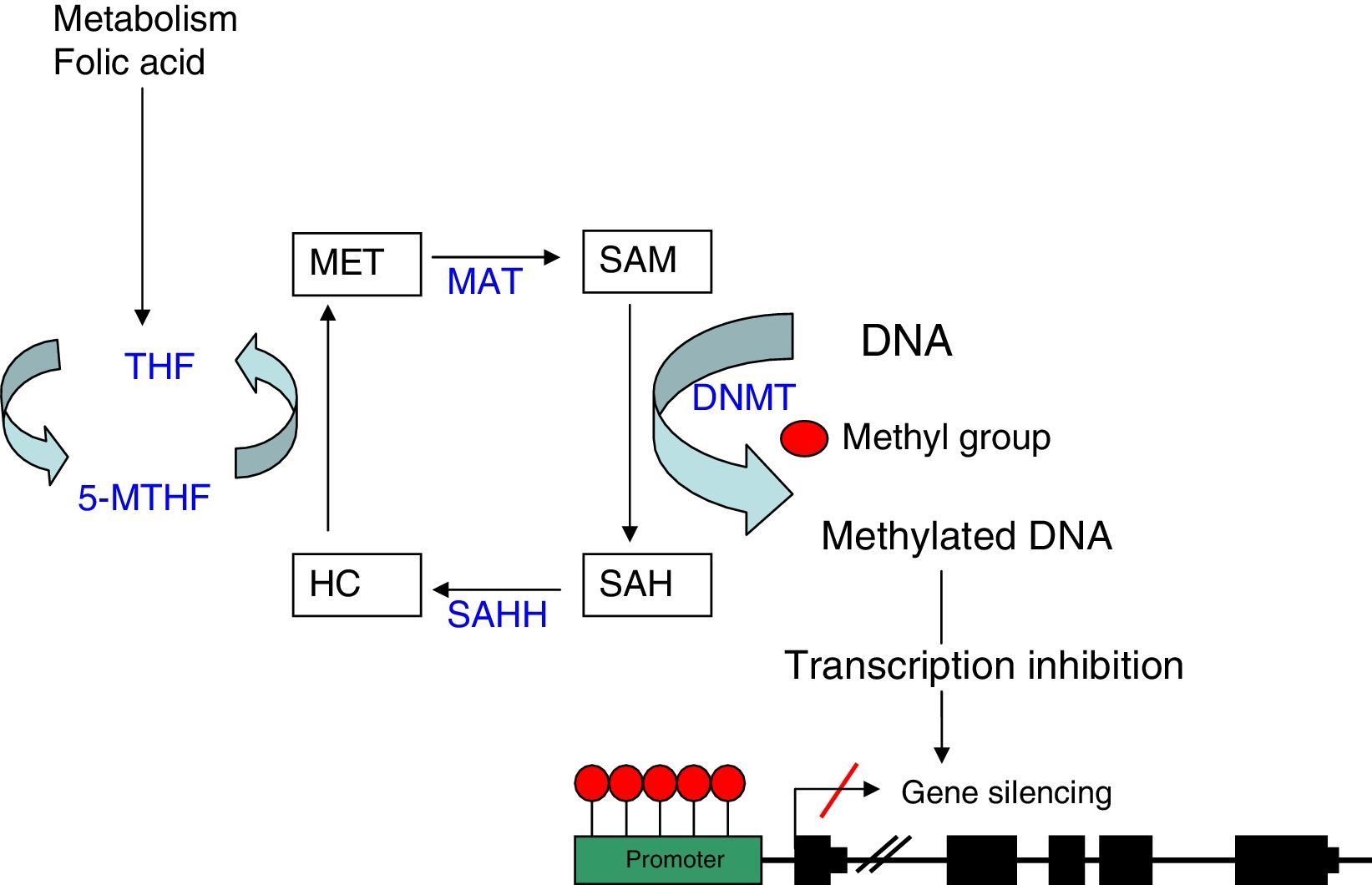
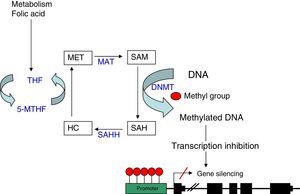
As explained before, the SAM molecule is the methyl group donor; this molecule becomes S-Adenosyl-l-homocysteine (SAH) after losing the methyl group. SAM is hydrolised to homocysteine and subsequently remethylated to methionine by 5-methyltetrahydrofolate cofactor (5-MTHF). Methionine is finally transformed into a SAM molecule by methionine adenosyltransferase (MAT) (Fig. 1). The potential of DNA methylation will depend on the ratio between SAM level and SAH in blood. The higher the ratio, the greater the potential of DNA methylation.8 We can therefore state that DNA methylation requires proper metabolism of homocysteine, methionine, other enzymes involved in this metabolic pathway, and such substances as folic acid and vitamin B12.9
DNA methylation: diagram of the metabolic pathway involved in DNA methylation. DNMT: DNA methyltransferases; HC: homocysteine, MAT: methionine adenosyltransferase; MET: methionine; SAH: S-adenosyl-l-homocysteine; SAHH: S-adenosyl-l-homocysteine hydrolase; SAM: S-adenosyl-l-methionine; THF: tetrahydrofolate; 5-MTHF: 5-methyltetrahydrofolate.
The link between alterations in epigenetic mechanisms and human disease has become an emerging line of research in recent years; evidence of such an association has been reported in several diseases, especially in the field of oncology. The first type of tumour to be associated with epigenetic regulatory mechanisms was colorectal cancer (CRC). A loss of overall methylation was initially observed in cancer cells of patients with CRC compared to those of healthy controls.10 At the same time, promoters of tumour suppressor genes were found to be methylated, leading to decreased expression of those genes.11 These findings supported the association between hypermethylation of tumour suppressor genes and disease development.
However, in other medical fields such as neurological diseases, the role disrupted DNA methylation plays in disease development is yet to be understood. In the specific case of multiple sclerosis (MS), epigenetic changes potentially involved in disease pathogenesis have recently been found, opening up a new and interesting line of research.
MS is considered the leading cause of severe neurological impairment in young and middle-aged adults. In Spain, prevalence studies report rates of around 80 cases per 100000 population. MS is a chronic disease causing inflammatory, demyelinating, neurodegenerative lesions to the central nervous system (CNS). Although its aetiology is still unknown, MS is believed to have an autoimmune, probably multifactorial origin associated with a number of genetic and environmental susceptibility factors. In view of the complexity of MS and the multiple aetiological mechanisms (both genetic and environmental) involved in MS pathogenesis, we may hypothesise that altered epigenetic regulation may be involved in MS.12,13
Risk factors and epigenetic changes in multiple sclerosisEpidemiological and familial aggregation studies suggest a genetic predisposition to MS. To date, however, the only locus consistently found to be linked to MS is the major histocompatibility complex (MHC). This predisposition has been linked to the DR2 haplotype (HLA-DRB1*1501-DQA1*0102-DQB1*0602), which is associated with a relative risk of MS of 4.14 The development of new techniques, such as polymorphism arrays, has led to the identification of new candidate genes located outside the MHC region. MS is a polygenic disease in which each of the associated genes present a different risk of developing the disease (normally low to moderate risk).15
The role of genetic susceptibility factors in MS is therefore clear; however, environmental factors causing epigenetic changes seem to be essential in MS development.16,17 We will now list the 3 environmental risk factors of MS described in the literature and analyse their impact on epigenetic regulation in MS and other diseases.18,19
Smoking and epigenetic mechanismsAs shown in multiple studies,20,21 smoking is one of the main environmental factors involved in progression of MS. In fact, smoking has been linked to increased frequency of relapses and a greater number of active brain lesions in MRI in patients with MS.22
A study analysing blood samples from adolescents whose mothers smoked during pregnancy found an association between prenatal exposure to tobacco and increased methylation of the brain derived neurotrophic factor (BDNF) promoter, which promotes differentiation and growth of new neurons.23 Suter et al.24 compared DNA methylation patterns in smokers and non-smokers using PCR tests. They found DNA methylation changes in 25 genes in non-smokers and in 438 genes in smokers.24 Epigenetic changes linked to smoking have also been found in studies of patients with cancer. In a study including patients with lung cancer, smokers exhibited hypermethylation of the CDKN2A, DAPK, and MGMT tumour suppressor genes.25 Likewise, a study of cervical cancer in women aged 15-19 years also observed hypermethylation of the CDKN2A gene in cervical epithelial cells of smokers.26
Vitamin D and epigenetic changesVitamin D deficiency is one of the most influential risk factors in the development of MS.27–29 Vitamin D is a powerful regulator of inflammatory and immunoregulatory response, acting on both innate and adaptive immunity.30
Although the mechanism by which vitamin D causes such changes remains unknown, the results of a study conducted by Joshi et al.31 suggest that it may be due to epigenetic changes. This study analyses the effects of 1,25(OH)2D3 (active form of vitamin D produced by the skin after exposure to ultraviolet light) on IL-17A production by CD4+ T-cells in humans. Researchers observed that 1,25(OH)2D3 directly inhibits IL-17, which is responsible for the transcription of proinflammatory cytokines by modifying histone deacetylase 2 (HDAC2) in the IL17A promoter region.31 Previous studies have shown that vitamin D is able to cause epigenetic changes, as in the case of CRC, where 1,25(OH)2D3 has been observed to induce expression of the gene coding for lysine-specific demethylase (JMJD3).32
Epstein–Barr virus (EBV) and epigenetic mechanismsTo date, several infectious agents have been serologically and pathologically linked to MS. One example is the study by Sundstrom et al.,33 which analyses evidence supporting the presence of a viral infection in the presymptomatic stage of MS. However, only EBV nuclear antigens displayed a direct pathological correlation with onset of MS.33,34
Thus, EBV infection has been linked to epigenetic changes in the infected cells. Several types of tumours have been associated with EBV infection due to hypermethylation of tumour suppressor gene promoters,35 as in the case of nasopharyngeal cancer or EBV-induced Hodgkin lymphoma, where hypermethylation of the promoter has been found to be caused by an increase in DNMT1, DNMT 3 a, and DNMT 3 b enzymes, mediated through viral protein LMP1.36 Epigenetic changes associated with EBV in MS are also linked to microRNA expression (miRNA). Expression of miR-142-3p in patients with MS has been linked to increased immune tolerance, while the expression of miRNA-155 is associated with greater T-cell differentiation and CNS inflammation.37
DNA methylation in multiple sclerosisThe precise pathophysiological mechanisms mediating between environmental risk factors and susceptibility to develop MS remain unknown.38 In this context, DNA methylation may provide new insights.
Inflammation and DNA methylationOver the past few years, numerous authors have linked the degree of methylation in specific genes to presence of MS. Kumagai et al.39 observed that the tyrosine phosphatase (SHP-1) promoter, a negative regulator of proinflammatory signalling, is hypomethylated in patients with MS compared to healthy controls. Methylation of the SHP-1 promoter results in decreased SHP-1 expression and consequently greater lymphocyte-mediated inflammatory activity.39
Janson et al.40 analysed CD4+ T cells in a series of patients with relapsing-remitting MS (RRMS). According to their findings, these patients displayed demethylation of FOXP3, a gene coding for scurfin (scurfin deficiency is associated with autoimmune diseases). FOXP3 demethylation may inhibit Th1 and Th2 cell differentiation while promoting regulatory T cells (Treg) and Th17 cells. Th1/Th2 and Treg/Th17 balance has an impact on the disease; imbalance may therefore result in new lesions or lesion repair. DNA methylation is precisely one of the factors regulating this balance.40,41
Other researchers report hypomethylation of the promoter of the gene coding for IL-17A, a proinflammatory cytokine secreted exclusively by activated T cells; hypomethylation has been associated with the development of autoimmune diseases and plays an essential role in MS pathogenesis.42
Demyelination and DNA methylationAccording to a study by Mastronardi et al.,43 the peptidyl arginine deiminase 2 (PAD-2) promoter is demethylated and PAD-2 is consequently overexpressed in the brain during white matter demyelination in patients with MS. PAD-2 leads to loss of myelin basic protein (MBP) stability as a consequence of the enzymatic conversion of arginine into citrulline. Citrullination causes the MBP to act as an antigen for T cells. PAD-2 promoter methylation in white matter is decreased by 25% in patients with MS compared to healthy controls. These changes are present in patients with MS but have not been observed in patients with such other neurological diseases as Alzheimer disease, Parkinson's disease, and Huntington disease.43
Neurodegeneration and DNA methylationTo date, no studies have specifically analysed the role of epigenetic mechanisms in neurodegeneration in patients with MS. Nevertheless, some studies have addressed the changes in DNA methylation occurring during neuronal death. Chestnut et al.44 analysed cells from patients with amyotrophic lateral sclerosis and observed that overexpression of the DNMT3a enzyme induces cell degeneration and death whereas DNMT3a inhibition protects neurons. In fact, DNA methylation regulates DNMT3a expression. The above suggests that such a mechanism may be involved in neurodegeneration in patients with MS.
ConclusionsMultiple sclerosis is a neurological disease with a considerable healthcare, social, and family burden. Despite recent advances in our understanding of MS, the exact aetiopathogenic mechanisms of the disease are yet to be determined and no curative treatment has been developed to date.
Epigenetic changes, such as DNA methylation, constitute a mechanism enabling environmental factors to affect individual gene expression. Epigenetics represents an interesting line of research in the medical field, especially in the case of diseases in which environmental risk factors may play a role, as in MS.
Despite the limited number of published studies of patients with MS, results encourage further research into this area. Currently available data point to a relationship between autoimmune diseases and regulation of DNA methylation in candidate genes, which are key to the development of MS.45,46 Despite mounting evidence of this association, further studies with larger patient and control groups should be conducted.47,48
Knowledge of the epigenetic changes involved in the pathogenesis of MS will help us understand the pathogenic mechanisms of the disease, paving the way for the identification of potential biomarkers and new therapeutic targets.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Iridoy Zulet M, Pulido Fontes L, Ayuso Blanco T, Lacruz Bescos F, Mendioroz Iriarte M. Modificaciones epigenéticas en neurología: alteraciones en la metilación del ADN en la esclerosis múltiple. Neurología. 2017;32:463–468.
This study has not been presented at the SEN's Annual Meeting or at any other meetings or congresses. This study has received no public or private funding.