Glioblastoma multiforme is the most common primary brain tumour, with the least favourable prognosis. Despite numerous studies and medical advances, it continues to be lethal, with an average life expectancy of 15 months after chemo-radiotherapy.
DevelopmentRecent research has addressed several factors associated with the diagnosis and prognosis of glioblastoma; one significant factor is tumour localisation, particularly the subventricular zone, which represents one of the most active neurogenic niches of the adult human brain. Glioblastomas in this area are generally more aggressive, resulting in unfavourable prognosis and a shorter life expectancy. Currently, the research into microRNAs (miRNA) has intensified, revealing different expression patterns under physiological and pathophysiological conditions. It has been reported that the expression levels of certain miRNAs, mainly those related to neurogenic processes, are dysregulated in oncogenic events, thus favouring gliomagenesis and greater tumour aggressiveness. This review discusses some of the most important miRNAs involved in subventricular neurogenic processes and their association with glioblastoma aggressiveness.
ConclusionsMiRNA regulation and function play an important role in the development and progression of glioblastoma; understanding the alterations of certain miRNAs involved in both differentiation and neural and glial maturation could help us to better understand the malignant characteristics of glioblastoma.
El glioblastoma multiforme es el tumor cerebral primario más común y con el pronóstico más desfavorable del sistema nervioso central. A pesar de los numerosos estudios y avances en medicina, este sigue siendo letal, con una esperanza de vida promedio de 15 meses posteriores a la quimiorradioterapia.
DesarrolloRecientemente, se han estudiado diversos factores asociados al diagnóstico y el pronóstico de pacientes con glioblastoma, como la localización tumoral, principalmente la zona subventricular; una de las áreas neurogénicas más activas del cerebro humano adulto. Los pacientes con glioblastoma asociados a esta zona en particular presentan generalmente una mayor agresividad, lo que resulta en un pronóstico desfavorable y una menor esperanza de vida. Actualmente, se ha profundizado en el estudio de los microARN, los cuales reflejan patrones de expresión distintos en condiciones fisiológicas o fisiopatológicas. Está reportado que los niveles de expresión de ciertos microARN, principalmente aquellos relacionados a procesos neurogénicos, se ven desregulados en eventos oncogénicos, favoreciendo así la gliomagénesis y la agresividad tumoral. En la presente revisión se discuten algunos de los microARN más importantes implicados en procesos neurogénicos de la zona subventricular y su asociación con la agresividad del glioblastoma.
ConclusionesLa regulación y función de los microARN desempeña un rol importante en el desarrollo y la progresión del glioblastoma; en consecuencia, la comprensión de las alteraciones de los microARN implicados en la diferenciación, así como en la maduración neural y glial, podrían ayudar a entender mejor las características malignas del glioblastoma.
Gliomas are a heterogeneous group of primary tumours of the central nervous system (CNS), and are classified by cell lineage (astrocytes, microglia, oligodendrocytes).1 Among this group of CNS tumours, glioblastoma multiforme (GBM) is the tumour with the poorest outcomes, with a mean life expectancy of approximately 15 months after diagnosis.2 GBM is currently considered an important public health issue due to its unfavourable prognosis and low survival rate.3 Despite advances in the use of molecular markers, such as hypermethylation of methylguanine methyltransferase and mutated or wild-type isocitrate dehydrogenase 1 (IDH1), no specific markers for early diagnosis of GBM have yet been identified.
In recent years, numerous studies have aimed to find biomarkers to improve the diagnosis and prognosis of GBM.4 However, the molecular patterns determining the aggressiveness of the tumour are not fully understood; therefore, it is essential to examine new factors associated with the disease. For example, the 2014 study by Liu et al.5 emphasises the role of microRNA (miRNA) in regulating the biogenesis of GBM. Studies of miRNAs in different types of cancer describe their importance as regulators of signalling pathways involved in the cell cycle, invasion, tumourigenesis, drug resistance, etc.6–8 It should also be noted that GBM is frequently associated with alterations in the expression of multiple miRNAs.9
Therefore, the dysregulation of miRNA profiles is a relevant area of research, as it may lead to the development of new treatment strategies in personalised medicine, particularly in the case of GBM involving the subventricular zone (SVZ), a neurogenic area. SVZ tumours have been shown to present properties favouring tumour aggressiveness, such as greater proliferative capacity, invasiveness, and recurrence10; these characteristics may be strongly related to the expression of certain miRNAs associated with neurogenesis in the SVZ.
This study addresses the function of miRNAs involved in subventricular neurogenesis and which are dysregulated in GBM, showing a strong association with the degree of malignancy.
One of the most representative characteristics of GBM is its heterogeneous cellular composition, including cells presenting little differentiation, such as neural stem cells (NSC), neural progenitor cells (NPC), and glial progenitor cells (GPC), and such well-differentiated glial cells as astrocytes, microglia, oligodendrocytes, and even neurons11,12; this composition gives the tumour pleomorphic, migratory, proliferative, and infiltrating capacities.13
Histopathological and molecular classification of glioblastoma multiformeBased on the cellular composition of the tumour, GBM may be classified as primary or secondary. Alternatively, some studies have evaluated the correlation between type of GBM and molecular alterations: primary GBM is mainly associated with mutation and overexpression of EGFR and MDM2, as well as deletion of p16 and loss of heterozygosity of chromosome 10q; and secondary GBM mainly features PDGFA/PDGFRa overexpression, loss of heterozygosity of chromosome 19q, and mutations in IDH1/2 and PT53.3 A new subclassification of GBM has been proposed, based on a meta-analysis of data from the Cancer Genome Atlas, and divides GBM into 4 subtypes: proneural, neural, mesenchymal, and classical, associated with alterations in the PDGFR, IDH1/2, NF1, and EGFR genes, respectively.14 Based on previous studies, the World Health Organization recently proposed a new classification of GBM: IDH-wildtype and IDH-mutant, combining histological and molecular markers and the status of the IDH1 gene.15
However, this classification cannot determine the behaviour or the prognosis of GBM; as a result, researchers are currently also studying the epigenetic regulation mechanisms of numerous miRNAs involved in such biological processes as gliomagenesis, cell renewal and proliferation, invasion, and drug resistance.16 It has also been suggested that these miRNAs may play an important role in tumour biology,17 regulating the expression of multiple targets, such as oncogenes, tumour suppressor genes, and growth factors, including K-RAS, CDK-4/6, BCL2, NOTCH, p53, EGFR, and VEGF, among others, which are involved in multiple biological functions as cell cycle control, cell differentiation and proliferation, infiltration, angiogenesis, and apoptosis.16,18
Tumour location: subventricular zoneAs discussed above, tumour location, another important aspect of the heterogeneity of these tumours, is highly relevant to the prognosis of GBM.19 The SVZ is one of the areas where adult neurogenesis occurs, and includes a structured niche of NSCs, NPCs, GPCs, glial cells, and vascular cells, all in constant communication. It has also been suggested that the origin and expansion of GBM occur in this neurogenic niche,20 due to the malignant transformation of NSCs and NPCs to glioblastoma stem cells (GSC), involved in the genesis and propagation of GBM,21 and the fact that a small population of tumour cells present characteristics of NSCs, such as multipotency, self-renewal, and expression of such cell markers as Nestin, Sox2, BMI1, and CD133.22 These characteristics support the hypothesis of a “tumour hierarchy” in which tumour stem cells are the only cells with a true capacity for self-renewal and multipotent differentiation.23
Several studies suggest that tumours involving the SVZ show a distinct growth pattern, with faster progression and more aggressive clinical behaviour.10,20,24 Thus, the development of GBM in the SVZ represents a potential predictor of unfavourable prognosis, with higher rates of recurrence and infiltration, as well as increased drug resistance. It has been suggested that presence of NSCs in this area is responsible for the malignancy of the tumour.20,25,26 As mentioned above, one factor determining tumour malignancy may be aberrant regulation of certain miRNAs, as they modulate important aspects of the behaviour and malignant capacities of GBM.27 Therefore, research into these components and their role in the aggressiveness of GBM in the SVZ is highly relevant.
MicroRNA: importance and activityMicroRNAs are short, non-coding RNA sequences of approximately 20-25 nucleotides in length, with the capacity to modulate post-transcriptional expression by binding to the 3-UTR region of target messenger RNA, inducing degradation of the messenger RNA and, in most cases, inhibiting the translation of proteins.28 As a result, miRNAs can modify the physiology of the cell, modulating such important processes as proliferation, differentiation, and survival.29
Given their extensive involvement in various cell processes, it has been suggested that miRNAs regulate up to one-third of human genes30; therefore, they undoubtedly also play a role in regulating the translation of oncogenes and tumour suppressor genes.31 For example, overexpression of miR-26a has been detected in approximately 12% of GBMs; this miRNA suppresses expression of PTEN and promotes gliomagenesis.32 Furthermore, miR-21, miR-17, and miR-221/222 are reported to be overexpressed in GBM; these molecules are mainly involved in anti-apoptotic functions and cell proliferation processes.16 The miRNAs miR-34a, miR-24, and miR-7 seem to inhibit tumour growth,8 whereas miR-195, miR-455-3p, and miR-10a are involved in resistance to temozolomide (TMZ) in GBM cells.33
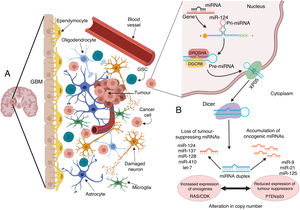
These studies highlight the importance of research into the role of miRNA in oncological processes: despite advances in our understanding of the regulation of pathogenic pathways in GBM, we also need to determine the epigenetic mechanisms regulated by miRNAs and understand how alterations in their expression can increase the malignant characteristics of the tumour (Fig. 1), especially with regard to anatomical localisation, mainly the SVZ, which confers properties favouring tumour growth.
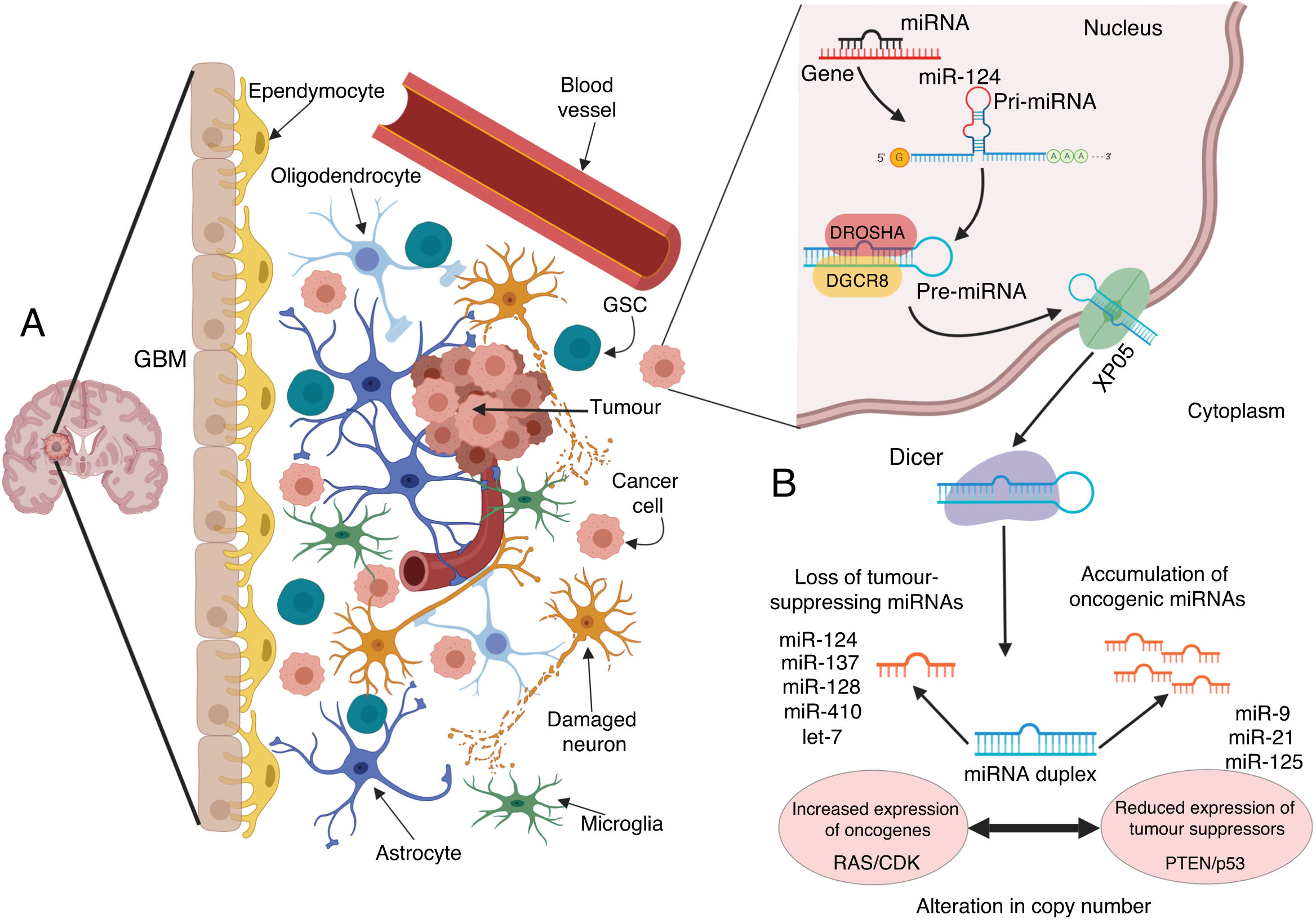
Microenvironment of glioblastoma and alterations in miRNA biogenesis, leading to growth and propagation of the tumour. A) GBM is a tumour composed of different cell types. This tumour niche presents both differentiated and undifferentiated cells that contribute to the onset, growth, and expansion of the tumour through multiple alterations to cancer cells, which modify the environment and the surrounding cells to their benefit; one of the most important alterations is the modification and transfer of molecules and genetic material such as miRNA. B) In such cancer cells as those of GBM, the biogenesis of miRNAs is altered by various processes that are commonly dysregulated by mutations and disorders in the processing, export, and maturation of miRNAs. In GBM, these modifications may alter the final number of copies of miRNA, leading to a reduction or loss of tumour suppressive miRNAs including miR-124, miR-137, miR-128, miR-410, and let-7; this translates to increased expression of oncogenes such as those belonging to the RAS and CDK families, among others. To the contrary, an increase may occur in levels of such oncogenic miRNAs as miR-9, miR-21, and miR-125, which silence such tumour suppressors as PTEN and p53.
Several transcriptome analyses performed to study miRNAs and GSCs have revealed that these cells cause tumours to present extraordinary aggressiveness, possibly due to dysregulation of various miRNAs that, among other effects, increase the tumour’s resistance to conventional therapy with TMZ34,35; likewise, other miRNAs that are usually suppressed in GBM, such as miR-124, miR-137, miR-128, miR-125b, and miR-181a/b, sensitise GSCs, reducing their resistance.36,37 On the other hand, it has been reported that various miRNAs specific to such brain areas as the SVZ modulate neurogenic functions including cell fate specification, regulation of cell development, and the maturation of NSCs and NPCs.38
The following section discusses some of the most widely reported miRNAs known to be involved in neurogenesis, which are dysregulated in GBM (Table 1).
MicroRNAs associated with neurogenesis in the subventricular zone and aggressiveness of glioblastoma multiforme.
| miRNA | Function in SVZ | References | Function in GBM | Expression in GBM | References |
|---|---|---|---|---|---|
| miR-124 | Neural differentiation, NPC proliferation, neurogenesis | 39–43 | Promotes GSC differentiation Reduces tumour stemness, proliferation, angiogenesis, invasiveness, and drug resistance | Reduced or absent | 44–47 |
| miR-137 | Neurogenesis, differentiation, maturation | 37,48,49 | Promotes GSC differentiation, cell-cycle arrest, and apoptosis | Reduced | 37,48,50–54 |
| Reduces tumour stemness, proliferation, migration, and invasiveness | |||||
| miR-9 | NSC differentiation, proliferation, neurogenesis | 55–57 | Promotes drug resistance | Increased | 58–62 |
| Reduces angiogenesis, proliferation, and invasiveness | |||||
| miR-128 | CNS development, neuronal maturation and migration, NPC proliferation | 63–65 | Promotes apoptosis | Reduced | 44,66–69 |
| Reduces GSC stemness, proliferation, angiogenesis, and resistance to chemo- and radiotherapy | |||||
| miR-21 | NSC development, maintenance, and proliferation | 70–72 | Promotes drug resistance, stemness, and proliferation of GSCs, angiogenesis, and invasiveness | Increased | 73–78 |
| Reduces apoptosis | |||||
| miR-410 | Neuronal and astroglial differentiation of NSCs | 79–84 | Reduces GSC proliferation, angiogenesis, invasiveness, and drug resistance | Reduced | 85–87 |
| Maturation and alteration of neuronal morphology | |||||
| miR-125 | Neurogenesis | 88–90 | Promotes invasiveness and proliferation | Increased | 36,91,92 |
| Differentiation and proliferation of NSCs and NPCs | Reduces or inhibits apoptosis | ||||
| Adult neurogenesis, neuronal maturation and differentiation | Promotes GSC differentiation | ||||
| Regulation of synaptic function | Reduces GSC stemness and proliferation | ||||
| let-7 | Reduces stemness and proliferation of NSCs and NPCs | 93–97 | Reduces the stemness of GSCs, proliferation and invasiveness, and chemo- and drug resistance; induces cell-cycle arrest; and reduces tumour growth | Reduced | 8,98–100 |
| Neuronal and astroglial differentiation | |||||
| Radial migration of NPCs |
This table summarises the normal and oncogenic function in GBM of the 8 miRNAs described in the article. It also shows the alterations in the expression of these miRNAs in GBM, with most being downregulated (miR-124, miR-137, miR-128, miR-410, and let-7) and others being upregulated (miR-9, miR-21, and miR-125). These alterations in regulation are mediated by various molecular components, leading to malignant properties.
CNS: central nervous system; GBM: glioblastoma multiforme; GSC: glioblastoma stem cell; NPC: neural progenitor cell; NSC: neural stem cell; SVZ: subventricular zone.
One of the most widely studied and most abundant miRNAs in the brain is miR-124,39 which is expressed in different stages of the NSC lineage in the SVZ: NSCs, transit-amplifying cells, neuroblasts, and mature neurons.40 MicroRNA-124 is involved in neurogenic events, such as neuronal fate specification; it has been reported that downregulation of miR-124 in progenitor cells in the SVZ may inhibit neuronal differentiation, whereas overexpression leads to neuronal determination41 and regulates the transition of transit-amplifying cells to neuroblasts,40 regulated by Sox9, PTBP1, and CDK6.42 However, miR-124 also downregulates Sox9, reducing the proliferation of precursor cells and stimulating neuronal differentiation by promoting expression of Tuj1 and MAP2.37,43 It has been reported that miR-124 is absent or that its expression is reduced in GBM, partly because the growth factors EGF, FGF, and PDGF promote its suppression.44 MicroRNA-124 is involved in tumour suppression and cell differentiation, inhibiting GSC proliferation by regulating SOS1,45 reducing angiogenesis, and increasing chemosensitivity to TMZ through negative regulation of oncogenes from the RAS family.46 Finally, overexpression of miR-124 can decrease the invasiveness of GSCs and promote their differentiation by silencing SNAI2, a gene that is overexpressed in glioma and that regulates stem cell function.47
Another of the most studied and most abundant miRNAs in the brain is miR-137.48 Its regulation of numerous target genes has been connected with neurogenic events in the SVZ, such as the neuronal differentiation of NSCs37; miR-137 also plays an important role in regulating the maturation of immature neurons.48,49 Levels of this miRNA are reduced in GBM, partly due to the fact that it undergoes hypermethylation in the context of tumours.48 Furthermore, as is also the case with miR-124, it is suppressed by the growth factors EGF, FGF, and PDGF.37 MicroRNA-137 has also been demonstrated to act as a tumour suppressor, decreasing the stemness of GSCs by reducing their capacity for self-renewal and inducing differentiation through silencing of LSD1, which, together with TLX, maintains mature NSCs in a self-renewable and undifferentiated state,50,51 partly through activation of the Wnt/β-catenin pathway52 and transcriptional repression of TLX target genes, such as p21 and PTEN, which stimulate the proliferation and self-renewal of NSCs.50 MicroRNA-137 is also involved in inhibiting proliferation, migration, and invasiveness and in inducing cell-cycle arrest in the G0/G1 stage through silencing of CDK6.37 Similarly, miR-137 is involved in the differentiation of GSCs and induction of apoptosis in GBM.37RTVP-1 has also been identified as a target gene for miR-137,53 which negatively regulates its expression in GBM, as RTVP-1 is associated with increased resistance to apoptosis.54 Its low expression suggests possible involvement in the maintenance of such tumour stem cells as GSCs; it is therefore one of the miRNAs most strongly associated with differentiation of neurons.
MicroRNA-9 has also been described in numerous studies into neurogenesis; this miRNA is specific to brain tissue, both at embryonic stages and in adults,55 and is strongly expressed in neurogenic areas at early ages, with extensive expression in the SVZ.56 Numerous transcriptional regulators have also been identified as direct targets of miR-9, including TLX, FOXG1, and HES1, which form a complex that regulates the proliferation and differentiation of NSCs and NPCs.57 Increased levels of miR-9 have recently been reported in GBM; this miRNA inhibits cell proliferation and migration and reduces invasive capacity and renewal.58 This tumour antagonism is associated with silencing of FOXP1,59 an oncogene that inhibits EGFR-dependent tumour growth, and whose expression is increased in GBM.60 Furthermore, it has been reported that inhibition of miR-9 promotes expression of VEGF-A, resulting in increased angiogenesis.61 However, other researchers report that miR-9 expression is upregulated in TMZ-resistant GBM, indicating that miR-9 reduces the expression of PTCH1, which promotes chemoresistance to TMZ in GBM; therefore, overexpression of miR-9 is associated with poor prognosis.62
MicroRNA-128 is abundant in brain tissue; expression of this miRNA has been related to the development of the CNS, contributing to cell diversity and plasticity and to neuronal maturation.63 Furthermore, miR-128 regulates the proliferation of NPCs in the SVZ64 and, together with PHF6, controls the migration of neurons through the cerebral cortex.65 It should be noted that miR-128 is one of the miRNAs presenting the most reduced expression in GBM44; this is related to the malignant potential of GBM, with the molecule acting as a tumour suppressor by reducing the stem capacity of GSC through inhibition of the epigenetic regulator PRC and the renewal factor BMI1, which act as oncogenes in GBM, potentiating the proliferation, maintenance, resistance, and renewal of NSCs and GSCs.66 MicroRNA-128 has also been shown to promote the sensitisation of GTCs to chemo- and radiotherapy through negative regulation of BMI1.67 Other researchers have reported that overexpression of miRNA-128 in GBM promotes a reduction in its capacity for growth, renewal, and cell invasion, in addition to its anti-proliferative, anti-angiogenic, and pro-apoptotic effects, through repression of such growth factors as RTK, EGF, and PDGFRa.68,69 On account of this, it has been suggested that loss of miR-128 expression is an early event in gliomagenesis,66 leading to decreased differentiation and increased GSC capacity for renewal and maintenance, resulting in more aggressive tumours.
MicroRNA-21 is expressed during the embryonic and neonatal development of the brain; furthermore, it is abundantly expressed in areas with large amounts of NGCs and GPCs, such as the SVZ.70 MicroRNA-21 is coexpressed with SOX2, which has been shown to participate in the proliferation and maintenance of NSCs, suggesting that miR-21 has similar functions.71 This microRNA also promotes signalling with such neurotrophins as NGF, which participate in controlling neuronal differentiation, and therefore neuronal development, survival, synaptic function, and plasticity.72 MicroRNA-21 is overexpressed in the majority of malignant gliomas, and is associated with poor prognosis. Overexpression of miR-21 is associated with angiogenesis, apoptosis, proliferation, and tumour invasion,73 as it favours increased expression of AKT and ERK, which upregulates the expression of HIF-1a and VEGF, promoting tumour angiogenesis.74 Inhibition of miR-21 in cultures of GBM results in activation of caspase-3 and caspase-7, leading to increased apoptotic cell death, as well as increased expression of BCL-2 and BAX; this suggests that miR-21 overexpression represents an anti-apoptotic factor, promoting chemoresistance.75,76 Such tumour suppressors as PDCD4, TPM1, and PTEN have also been identified as targets silenced by miR-21,77 triggering an increase in proliferation and invasion. A 2016 study by Hermansen et al.78 suggests that overexpression of miR-21 begins in the early stages of tumour development, increasing the capacity of GSCs for self-renewal and proliferation.
Another important molecule is miR-410, which is extensively expressed in CNS cells, where it was first described, and limited to the developing CNS79; miR-410 belongs to the miR–379-410 group of miRNAs, which are involved in numerous processes of neurological development, regulating neuronal differentiation, proliferation, and migration.80,81 Specifically, miR-410 is expressed in the SVZ; it is also transiently expressed in mesenchymal stem cells. MicroRNA-410 is also known to be downregulated by the Noggin protein, a transcription factor synthesised by ependymal cells that antagonises BMP-4, synthesised by astrocytes, enabling differentiation of the new cells towards neuronal rather than glial lineage.82 Downregulation of miR-410 is reported to promote neuronal differentiation and decrease astroglial differentiation of NSCs; in turn, this miRNA is also involved in maturation and in the alteration of neuronal morphology.83,84 Expression of miR-410 is markedly downregulated in GBM, compared to less malignant gliomas and healthy brains.85 A study by Chen et al.86 demonstrated that expression of miR-410 can significantly inhibit proliferation and reduce the invasive capacity of glioma cells, acting as a tumour suppressor; inhibition of miR-410 had the opposite effect. Likewise, these researchers showed an inverse correlation between the expression of miR-410 and MET, a protein targeted by miR-410.86 MET has also been connected to mesenchymal GBM and metastatic cancer, with high levels of expression in different tumours; it promotes evasion of apoptosis, increased cell proliferation, invasiveness, angiogenesis, and drug resistance, providing an escape route for tumours to reappear in more aggressive forms.87
Another miRNA that is extensively expressed in the brain tissue is miR-125b; this molecule is overexpressed in NSCs and neuroblasts of the SVZ during neurogenesis, promoting their proliferation and differentiation,88 and has also been identified as a marker of adult neurogenesis and neuronal maturation, as it is expressed by mature neurons.89 This miRNA regulates the lineage commitment of NSCs, promoting neuronal and astroglial differentiation; furthermore, it contributes to regulating synaptic function, as the majority of its target genes are related to this function.90 The role of the miR-125 family in GBM is somewhat controversial, with some studies suggesting that miR-125b expression is increased in glioma36 and is directly related to the degree of malignancy and general survival; according to these data, miR-125b would act as an oncogene, repressing tumour suppressors and apoptotic signalling pathways such as the p53 and p38 MAPK pathways, thereby promoting invasiveness and proliferation and inhibiting apoptosis in GBM cells.91 On the other hand, other researchers report that miR-125b expression is downregulated in GBM and suggest that the molecule acts as a tumour suppressor, promoting differentiation and reducing proliferation of GSCs through negative regulation of the TAZ mediator and its target oncogenes, which include CTGF and survivin, involved in tumour formation and progression.92
Finally, the let-7 miRNA is very abundant in NSCs and immature neurons,93 and participates actively in several processes, including the radial migration of neuroblasts94 and the regulation of cell fate specification for NSCs and NPCs, playing a key role in lineage decision-making between neural and glial development.95 Overexpression of let-7 reduces the self-renewal and proliferation of NSCs, promoting neuronal and astroglial differentiation; reduced expression of let-7 has the opposite effect.96 This reduction or inhibition of let-7 may be regulated by the RNA-binding protein Lin28, promoting greater expression of let-7’s target genes c-Myc, HMGA2, RAS, CCND1, and TLX; these genes are positively involved in the proliferation of NSCs and NPCs.97 Let-7 has also been identified as a tumour suppressor in gliomas, and its levels are generally reduced in GBM.8 Let-7 may reduce the aggressiveness of GBM, as it is reported to present anti-proliferative and anti-migratory properties due to silencing of such oncogenes as MYC, N-RAS, and K-RAS, which are involved in GBM proliferation, invasion, and survival.98 The low expression of members of the let-7 family in various types of cancer may be explained by activation of Lin28A and Lin28B; this phenomenon has been associated with deficient response to chemo- and radiotherapy.99 Consequently, increased expression of let-7 in GBM cells may sensitise them to targeted therapies. Furthermore, a study published in 2016 by Song et al.100 reports that overexpression of let-7b inhibits tumour growth and GSC stemness in vitro.
Bioinformatic tools for microRNA analysisComputational prediction has been highly valuable in identifying miRNA targets, as this is the most important action in clarifying their regulatory functions.101 Numerous programs and algorithms have been developed to conduct large-scale multivariate analysis of miRNAs and to identify miRNAs and their levels of expression, to predict their precursors and targets, to visualise their functions, to establish miRNA transcription networks, etc.102,103
Such bioinformatic tools as miRNet enable easy identification of the targets and signalling pathways of different miRNAs and their association with more specific cellular functions.104 Even when the target transcripts of a specific miRNA have been identified, understanding or visualising their effects is not straightforward; furthermore, miRNAs may have hundreds of distinct targets and can therefore affect a wide variety of processes.
In turn, the generation and use of different specialised databases supports the study and characterisation of biological systems; for instance, miRBase contains a registry of more than 2600 mature human miRNA sequences.105 Of these, the miRTarBase106 database includes reports of at least 118 distinct miRNAs together with their target genes for GBM.
A large quantity of information has been generated in recent years on new miRNA-based diagnostic and prognostic biomarkers, both in normal and in pathological cell processes.107 However, the regulation mechanisms of these miRNAs and their target genes, and the physiological implications of these processes, are not well understood.102 Therefore, the use of databases combined with bioinformatic tools may facilitate large-scale compilation and analysis, providing the information needed to construct networks of miRNA-target interactions and to better understand these mechanisms and processes. This article used the online miRNet tool104,108 and the Kyoto Encyclopedia of Genes and Genomes (KEGG)109–111 and miRBase112–115 databases, as well as the literature on the subject, to construct maps of miRNA-target interactions for miRNAs expressed in the SVZ and dysregulated in GBM, in order to draw conclusions on their association with GBM aggressiveness and neurogenesis in the SVZ.
In silico analysis of microRNA markersIt is important to identify the target genes of miRNAs and their interactions with signalling pathways involved in different cell processes with a view to clarifying the functions of miRNAs; to that end, we performed an in silico analysis using the miRNet online platform. This tool uses the miRBase database for miRNA, the National Center for Biotechnology Information database for target genes, and the KEGG database to visualise signalling pathways. We selected the 8 miRNAs described above to generate a map or network of interactions between miRNAs and their respective target genes, enabling us to visualise the signal pathways that are potentially altered in GBM. The analysis was divided into 2 parts: the first shows the 4 miRNAs most commonly reported in glioblastoma in an interaction diagram created with the KEGG Mapper online tool, part of the KEGG platform (Fig. 2). Subsequently, we analysed the interaction of the 8 miRNAs described; for this analysis, we used the online miRNet platform to generate a map of miRNA-gene interactions including all 8 miRNAs and the genes regulated by them in GBM (Fig. 3).
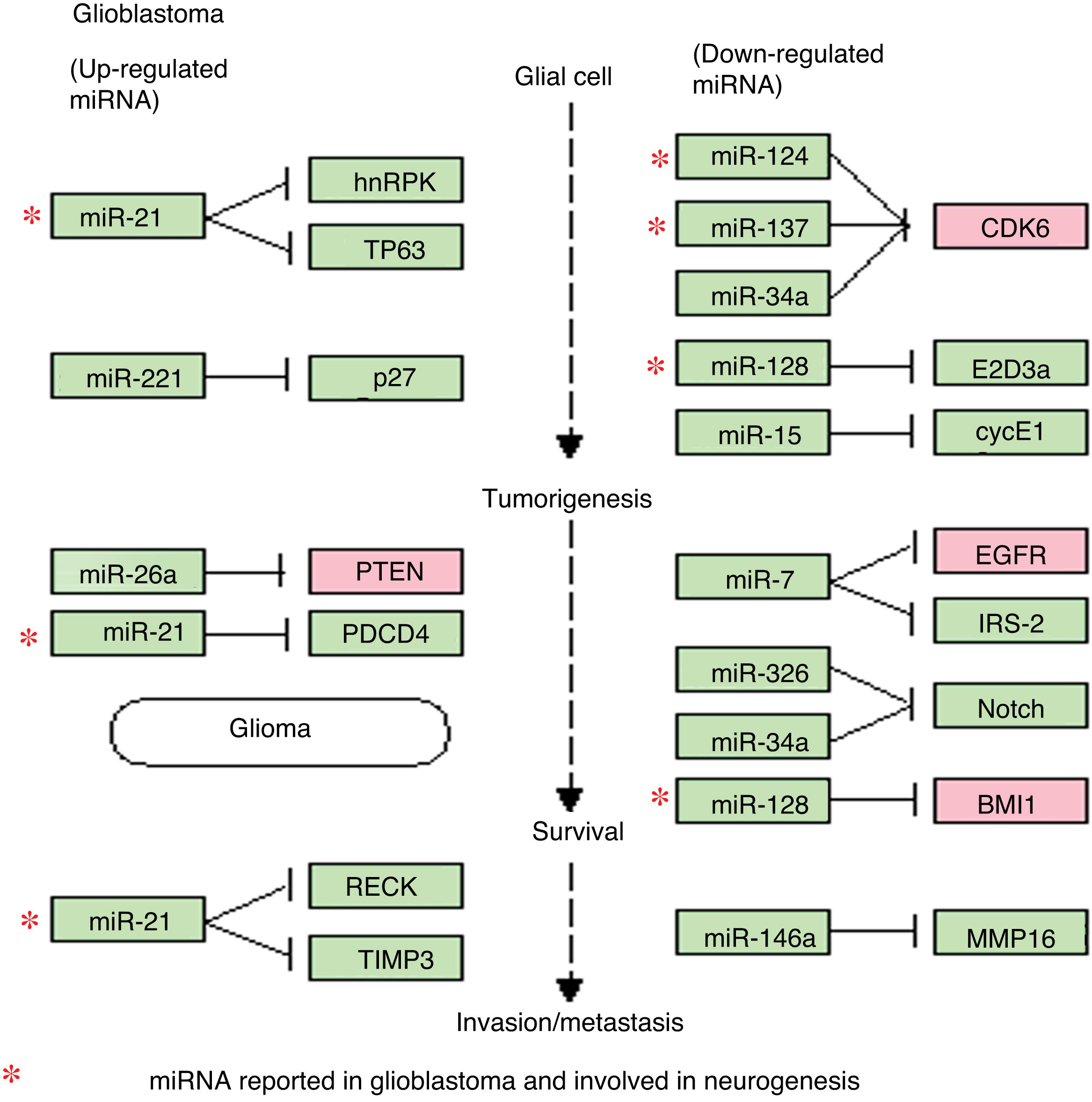
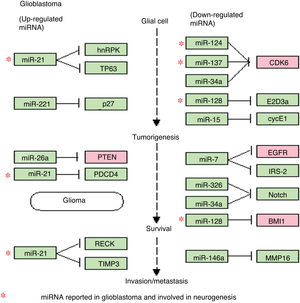
Regulation of microRNAs and their target genes in glioblastoma. This diagram of miRNAs in glioblastoma, generated with the KEGG online platform, shows the dysregulated miRNAs most frequently observed in glioblastoma and their main target genes. This diagram includes 4 of the 8 miRNAs described in the article (miR-21, miR-124, miR-137, and miR-128), all of which are involved in various processes of gliomagenesis and GBM aggressiveness. The diagram demonstrates the relevance of miR-21 (upregulated in GBM), which inhibits apoptosis and increases drug resistance, and is involved in the maintenance and proliferation of GSCs. It also shows the roles of miR-124, miR-137, and miR-128 (downregulated in GBM), which are involved in cell differentiation, induction of apoptosis, and cell-cycle arrest and the induction of chemosensitivity.
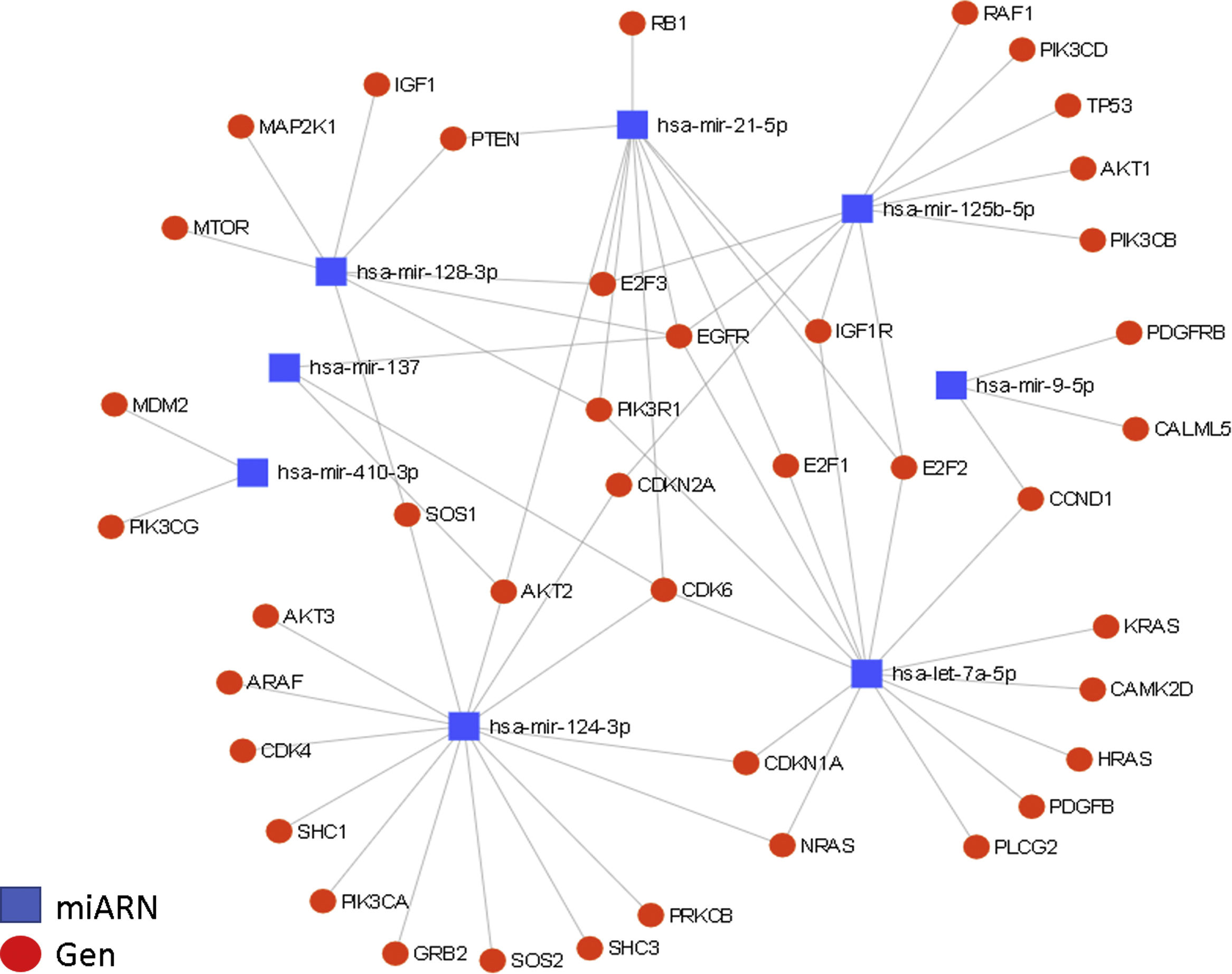
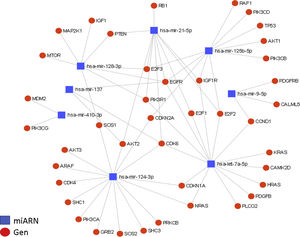
Interactions between miRNAs and their target genes (diagram generated with the miRNet online platform). A total of 36 target genes are shown for the 8 miRNAs in the context of glioma, grouped in nodes for each miRNA-gene interaction, with an affinity value of 8.31e–11.
The regulation of the EGFR and CDK6 genes is particularly relevant. EGFR is regulated by at least 5 of the miRNAs described in this article (miR-21, miR-125, miR-137, miR-128, and let-7); this growth factor is generally overexpressed in GBM and is partially responsible for the increased cellular proliferation and evasion of apoptosis. The CDK6 gene is regulated by 4 miRNAs (miR-124, miR-137, miR-21, and let-7); it is also overexpressed in GBM, and is associated with increased tumour progression, migration, and invasiveness.
The diagram of miRNA regulation shows the importance of some of these miRNAs, such as miR-21 (overexpressed in GBM), which is involved in malignancy of GBM, as well as the negative regulation of such other miRNAs as miR-124, miR-137, and miR-128 (underexpressed in GBM), which act as tumour suppressors and whose anti-tumour properties are greatly diminished if they are absent or expressed at low levels, resulting in an increase in the malignant potential of GBM. We also observed that several genes are regulated by multiple miRNAs: EGFR and CDK6 are regulated, respectively, by 5 and 4 of the miRNAs described. Both genes are overexpressed in GBM and play a significant role in malignant processes, directly influencing tumour progression, as both are related and potentiate such oncogenic events as tumorigenesis, growth, proliferation, evasion of apoptosis, and chemoresistance in GBM.
Therapeutic applications of microRNAs in glioblastoma multiformeDue to their capacity to interact with different targets and signalling pathways, miRNAs represent an important therapeutic target, with 2 possible approaches: either reducing or stimulating their expression, using different strategies, due to their capacity to act either as oncogenes or as tumour suppressors, which has led to the development of miRNA-based therapies and treatments targeted against these molecules in the treatment of GBM.8,116 This may be a plausible strategy due to the relative simplicity of obtaining genomic information and a personalised miRNA profile, given the continuing development and increasing use of microarrays and computational and bioinformatic tools.117,118 Thus, profiling miRNA expression would provide a clinical basis to improve diagnosis, enabling us to distinguish between tumours of the same histopathological grade, to more accurately establish prognosis in terms of general survival, and to estimate treatment response in individual patients.119
ConclusionsGBM is an important medical and social problem worldwide, as it presents the greatest prevalence and poorest prognosis of all CNS tumours; its aetiology is not fully understood. It has been suggested that GBM can arise from mutations in stem cells or from less malignant tumours, such as astrocytic tumours. However, although aetiology may be multifactorial, one of the most important aspects in its progression and prognosis is localisation, with the SVZ, the main neurogenic niche with abundant NSCs with multipotent and proliferative capabilities, being the region where tumour development is most aggressive.
Despite significant advances in the study of this disease, we currently only understand some of the molecular factors associated with GBM, with miRNAs being the main example; miRNAs participate in numerous physiological and pathological processes. Profiles of miRNA expression are dysregulated in GBM, influencing both the genesis and the development of the tumour; therefore, these molecules constitute interesting candidates for the study of the pathological processes underlying GBM.
Through a literature review and consultation of databases, this article characterises the relationship between different miRNAs and the development of GBM. The most relevant neurogenic miRNAs are miR-124, miR-137, miR-9, miR-128, miR-21, miR-410, miR-125, and let-7, all of which serve numerous functions in stem cells in the SVZ, influencing maintenance, differentiation, and fate specification of NSCs, the proliferation and migration of NPCs and GPCs, and other functions.
The importance of miRNAs in a neurogenic niche like the SVZ is evident, as are their implications in neural and glial differentiation and maturation. Alterations in miRNA profiles play an important role in cancer, influencing the development and progression of GBM and the maintenance of GSCs, inhibiting neural and glial differentiation, and increasing proliferation, angiogenesis, and drug resistance; all of these characteristics are observed in tumours of the SVZ. Therefore, better understanding of alterations in the function and regulation of these miRNAs may clarify the field of possible causes of aggressiveness in GBM involving the SVZ, and generate important diagnostic and prognostic markers for patients with GBM.
Literature reviewThe literature review was performed using the National Center for Biotechnology Information database and the PubMed search engine, with the following search terms: microRNA, miRNA, neurogenesis, differentiation, glioma, glioblastoma, bioinformatics, therapeutic. From the search results, we selected the articles to review and summarised the data reported.
Conflicts of interestThe authors have no conflicts of interest to declare.
We are grateful to the Mexican National Council of Science and Technology for the grants provided to L. J. Reséndiz-Castillo (grant no. 488270) to complete a masters degree and to E. E. Reza-Zaldivar (grant no. 487713) for doctoral study.
Please cite this article as: Reséndiz-Castillo LJ, Minjarez B, Reza-Zaldívar EE, Hernández-Sapiéns MA, Gutiérrez-Mercado YK, Canales-Aguirre AA. Efecto de la alteración de los niveles de expresión de microARN neurogénicos y su implicación en la agresividad de glioblastomas localizados en la región paraventricular. Neurología. 2022;37:781–793.