To evaluate the possibility of determining the genetic profile of primary malignant tumors of the breast from specimens obtained by ultrasound-guided percutaneous biopsies during the diagnostic imaging workup.
Material and methodsThis is a retrospective study in 13 consecutive patients diagnosed with invasive breast cancer by B-mode ultrasound-guided 12 G core needle biopsy. After clinical indication, the pathologist decided whether the paraffin block specimens seemed suitable (on the basis of tumor size, validity of the sample, and percentage of tumor cells) before sending them for genetic analysis with the MammaPrint® platform.
ResultsThe size of the tumors on ultrasound ranged from 0.6cm to 5cm. In 11 patients the preserved specimen was considered valid and suitable for use in determining the genetic profile. In 1 patient (with a 1cm tumor) the pathologist decided that it was necessary to repeat the core biopsy to obtain additional samples. In 1 patient (with a 5cm tumor) the specimen was not considered valid by the genetic laboratory. The percentage of tumor cells in the samples ranged from 60% to 70%. In 11/13 cases (84.62%) it was possible to do the genetic analysis on the previously diagnosed samples.
ConclusionIn most cases, regardless of tumor size, it is possible to obtain the genetic profile from tissue specimens obtained with ultrasound-guided 12 G core biopsy preserved in paraffin blocks.
Evaluar la posibilidad de obtener el perfil genético de los tumores primarios malignos de la mama a partir de las muestras obtenidas mediante la biopsia percutánea ecoguiada realizada durante el proceso diagnóstico.
Material y métodosEstudio retrospectivo sobre 13 pacientes consecutivas diagnosticadas de cáncer infiltrante de mama mediante biopsia ecoguiada (Modo B) con aguja gruesa (BAG) de calibre 12 G. Tras indicación clínica, el anatomopatólogo determinó, sobre los bloques de parafina, la aparente idoneidad de las muestras, antes de enviarlas al laboratorio para análisis genético con la plataforma MammaPrint®. Se han evaluado los siguientes aspectos: tamaño tumoral, validez de la muestra y porcentaje de células tumorales.
ResultadosEl tamaño ecográfico tumoral osciló entre 0,6cm y 5cm. En 11 pacientes la muestra conservada se consideró “válida” y apta para la determinación del perfil genético. En una paciente (tamaño tumoral de 1cm) y a juicio del anatomopatólogo, hubo que repetir la BAG para obtener muestras adicionales. En otra paciente (tamaño tumoral de 5cm), la muestra no fue considerada “válida” por el laboratorio genético. El porcentaje de células tumorales, entre las muestras válidas, osciló entre el 60% y 70%. En 11 de 13 (84,62%) casos fue posible el análisis genético a partir de las muestras diagnósticas previas.
ConclusiónResulta posible obtener el perfil genético en la BAG ecoguiada con agujas de 12 G, a partir de las muestras diagnósticas conservadas en bloques de parafina, en la mayoría de los casos, independientemente del tamaño tumoral.
Breast cancer is a heterogeneous disease; therefore, knowledge of the so-called profile (“fingerprint”, “signature”) of tumor gene expression (genotype) has become an important tool when it comes to establishing prognosis and planning therapy in patients affected by breast cancer.1–5
This information is usually obtained through surgical tumor samples. However, several situations occur in which it is necessary to obtain such information non-surgically (especially if implementing neoadjuvant therapy is being considered) through breast percutaneous biopsy (PB).
There are not a lot of specific bibliographic references regarding the exclusive use of the paraffin blocks resulting from diagnostic PB; therefore, our goal is to determine whether diagnostic PB samples are adequate to evaluate the tumor genetic profile through the genetic platform MammaPrint®.
Material and methodsRetrospective study of 13 consecutive patients between January 2012 and September 2015. Inclusion criterion: diagnosis of infiltrating breast cancer through PB later confirmed through surgery, with a clinical indication to determine the genetic signature from the samples obtained during the diagnostic process. No exclusion criteria have been considered.
The protocols established in our centers to access information in the medical histories have been followed in order to be able to publish it. All patients gave their written informed consent for the determination of the genetic profile. It was not necessary to request authorization from the Hospital Ethics Committee since no resource or procedures different from those already registered in everyday practice were used.
The percutaneous biopsies corresponded to a B-mode ultrasound-guided biopsy with a thick needle on masses suspicious of malignancy and were conducted by the same radiologist using a 12 G Trucut needles (Magnum®, BARD). The number of samples per patient ranged from 3 to 4 and was obtained by selecting the “long” (22mm) advance and that of different areas of the lesion following our usual protocol.
After the clinical request, the pathologist would review the paraffin blocks and determine the suitability of the samples, before referring them to perform the genetic workup using the MammaPrint® software (Agendia Inc., Amsterdam, The Netherlands). The sample was considered valid whenever the genetics lab would say so, considering the specimen suitable in the presence of at least 30% tumor cells.6 After the genetic analysis, the sample was returned to the anatomical pathological lab.
The genetic analysis classified the patients into two groups, “high” or “low” based on the level of risk of developing recurrence/metastatic disease.
The following aspects of each case were evaluated: tumor size, sample validity and percentage of tumor cells.
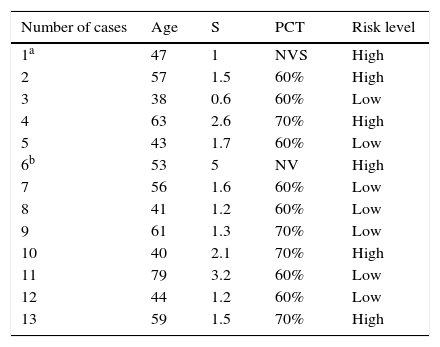
ResultsTable 1 shows the list of cases with the patients’ ages, tumor size, percentage of tumor cells in the samples analyzed and level of risk obtained in the analysis.
Age, tumor size, percentage of tumor cells and risk level.
| Number of cases | Age | S | PCT | Risk level |
|---|---|---|---|---|
| 1a | 47 | 1 | NVS | High |
| 2 | 57 | 1.5 | 60% | High |
| 3 | 38 | 0.6 | 60% | Low |
| 4 | 63 | 2.6 | 70% | High |
| 5 | 43 | 1.7 | 60% | Low |
| 6b | 53 | 5 | NV | High |
| 7 | 56 | 1.6 | 60% | Low |
| 8 | 41 | 1.2 | 60% | Low |
| 9 | 61 | 1.3 | 70% | Low |
| 10 | 40 | 2.1 | 70% | High |
| 11 | 79 | 3.2 | 60% | Low |
| 12 | 44 | 1.2 | 60% | Low |
| 13 | 59 | 1.5 | 70% | High |
NVS, non-valid sample; PCT, percentage of tumor cells notified by the genetics laboratory; S, size of the tumor on which the biopsy was performed (in cm).
The patients’ ages ranged from 38 to 79 years, with an average age of 52 years. Tumor size ranged from 0.6cm to 5cm, with an average size of 1.88cm.
In one of the cases (case #1), the pathologist determined that there was not enough neoplastic volume, and the biopsy had to be repeated, and then a valid sample was obtained. In the remaining cases, the pathologist considered the previous sample as valid, and it was sent to the laboratory without having to re-take new samples. However, in one case (case #6), the MammaPrint® lab considered the sample not valid.
In 11 of the 13 cases (84.62%), the samples were considered valid and it was possible to obtain the genotype of the tumor.
The percentage of tumor cells notified by the genetics lab ranged from 60% to 70%. Of the 11 valid samples, 7 contained 60% of tumor cells and 4 samples contained 70%, with a percentage of 60%.
In 7 cases, a “low” risk genetic profile was obtained (Fig. 1), and in 4 a “high” risk genetic profile was obtained (Fig. 2). The two cases in which it was not possible to perform the genetic analysis from the diagnostic PB were tumors with high-risk genetic profiles, and the result was obtained by re-taking the samples (case #1) or from a surgical sample (case #6).
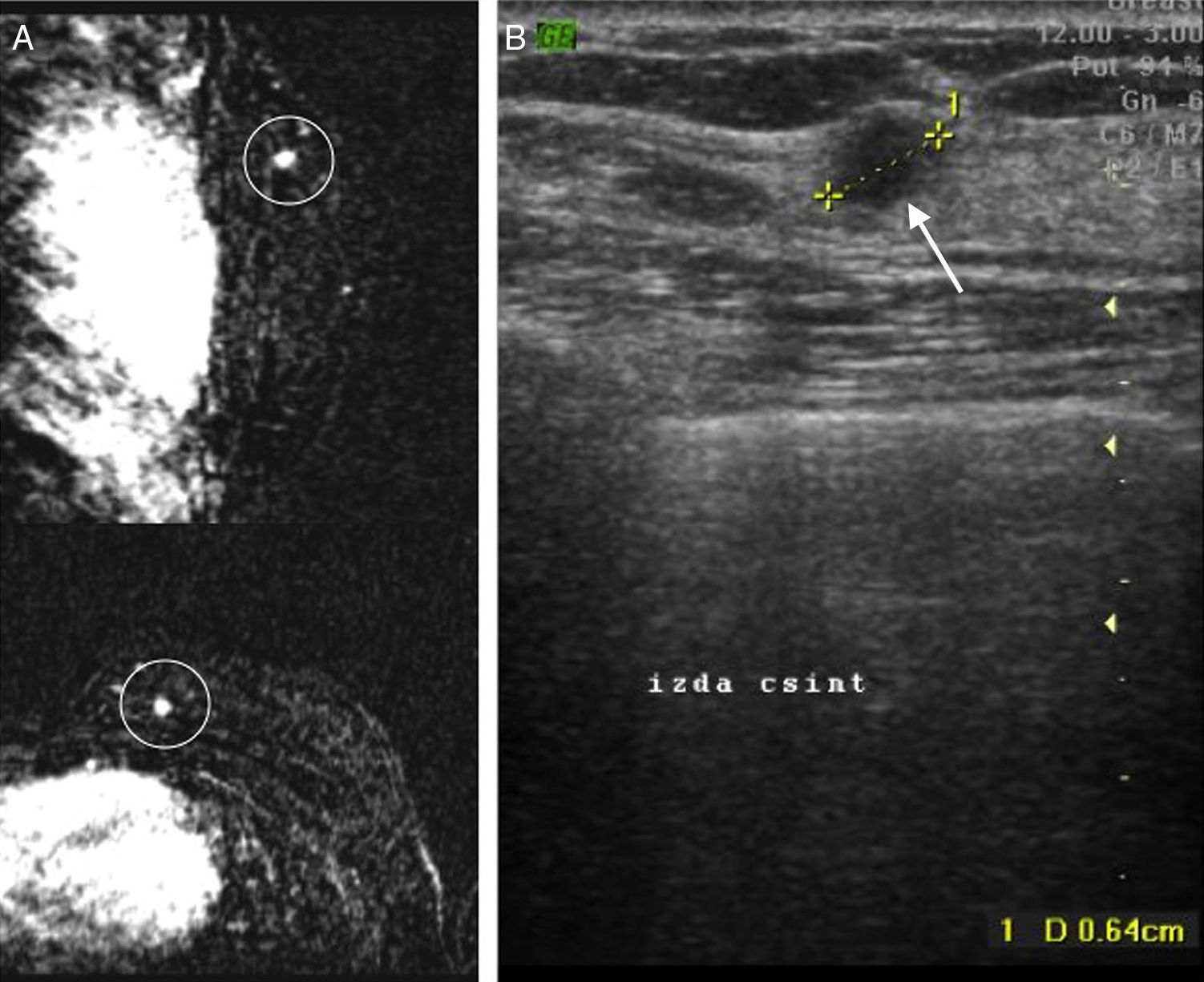
Case #3: 38-year-old patient with a history of right breast cancer. In the routine control conducted through MRIs (A), and in the contralateral breast (left), one 0.6cm mass suspicious of malignancy (circle) and corresponding ecographically (B) to the finding marked with the arrow. Determination through MammaPrint®: low risk.
The appearance of genetic platforms has been a significant endorsement to early detection practices, since it has been demonstrated that cases detected early show a more favorable genetic signature.7 But it also allows us to implement personalized therapies and thus obtain better, more efficient results, by selecting the patients that can benefit, or not, from a systemic pharmacologic therapy, as the type of systemic pharmacologic therapy used.1,4,5,8
Our work group selected MammaPrint® from several platforms available in the market. According to some authors9 this platform,3 the first to be approved by the Food and Drug Administration (FDA), is the most efficient and adequate one to establish prognosis and choose therapy.
At the beginning determining the genotype of breast tumors using the MammaPrint® platform required fresh surgical samples. However two events were a remarkable breakthrough on this regard. On one hand, the works by several authors10–13 that optimized modalities and techniques to obtain the genetic expression from very small samples, and on the other, the favorable results4,12,14,15 to do so from paraffin blocks, with results very similar to those obtained in surgical samples.15–17
Soon appeared publications that highlighted the possibilities of PB with the use of the aforementioned platform, with very few bibliographic references that would clearly specify the methodology followed when taking the PB, though the goal in all of them is to obtain additional samples, after knowing the diagnosis of malignancy.
In the year 2008 there appeared bibliographic references18 about the use of ultrasound-guided PB in 50 patients, with 14 G needles, with 70% of additional (fresh) samples suitable for analysis with MammaPrint®.
In the year 2011 there appeared bibliographic references19 from samples obtained through ultrasound-guided vacuum-assisted biopsy (VAB) in 67 patients (in several periods), with 11 G needles, and an average percentage of 73% of samples (both fresh and in paraffin blocks) suitable for analysis. As in the case of the work by Mayordomo et al.18 the samples were obtained additionally.
In the same year, Uribe et al.20 published a series of 16 patients with malignant tumors (previously diagnosed) of up to 2cm in size, analyzed using the MammaPrint® from additional surgical samples collected with “punch” and PB, both by the surgeon (in some cases) and the pathologist (other cases), without specifying other methodological details. The percentage of valid samples notified was 53.75% that the authors attribute to the low effectiveness of the “punch” or to tissue fibrosis associated to the small size of the tumors studied.
Since we have not found any bibliographic references about experiences of a methodology similar to ours, with the exclusive use of samples on which the diagnosis of malignancy was attained to determine the tumor genetic signature, our percentage of initial valid samples (84.62%) cannot be compared to that achieved by other authors.
It would be reasonable to assume that the greatest percentage of tumor cells and consequently, the greater probability of obtaining valid samples would depend on the following factors: the size of the tumor, the gauge of the needle, the number of samples and the type of sampling with the needle during the taking of the sample (or the element operator).
In all cases we performed the ultrasound-guided PB by selecting the “long” mode of needle cut because we believe that this ensures obtaining more tumor cells.
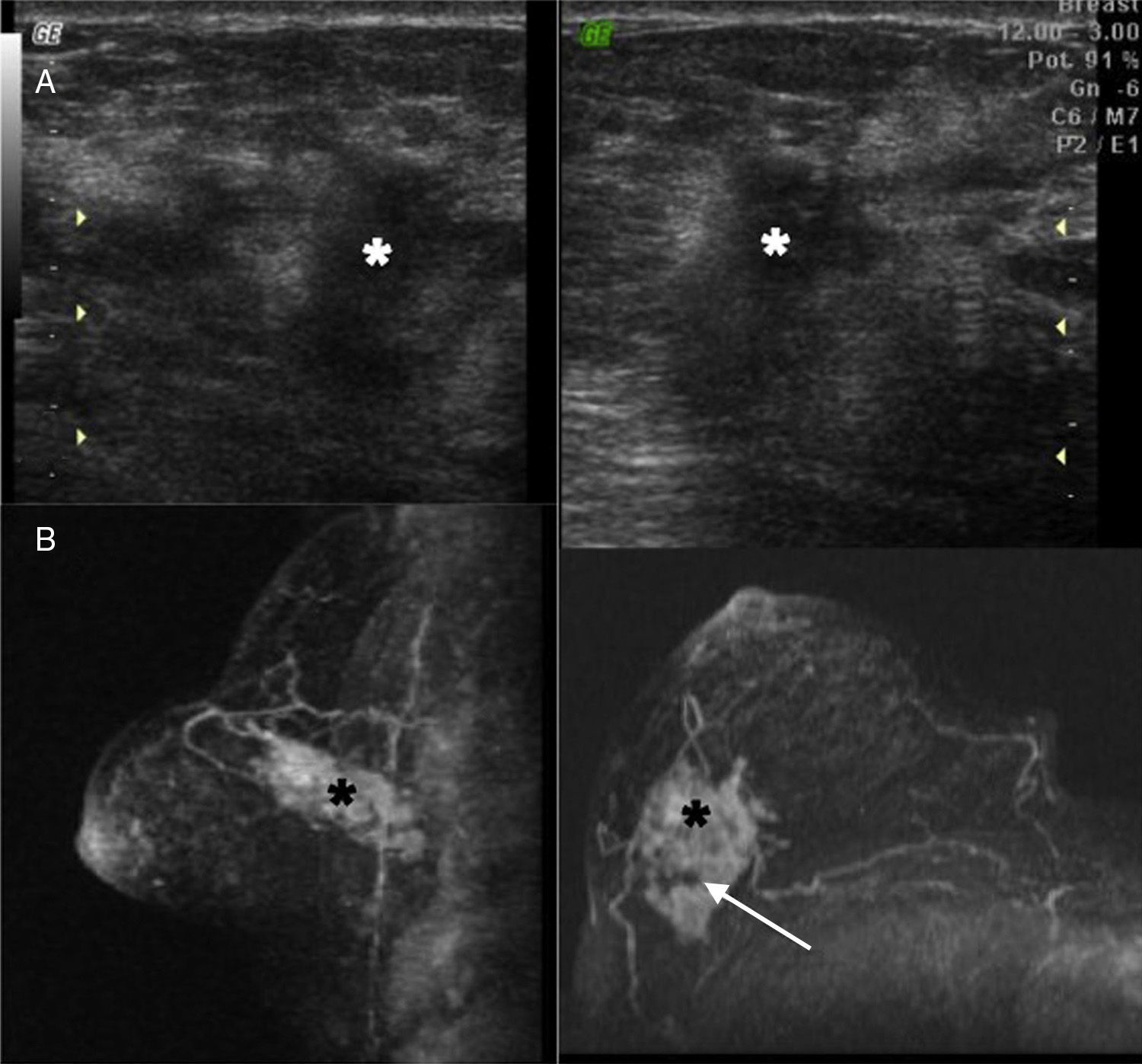
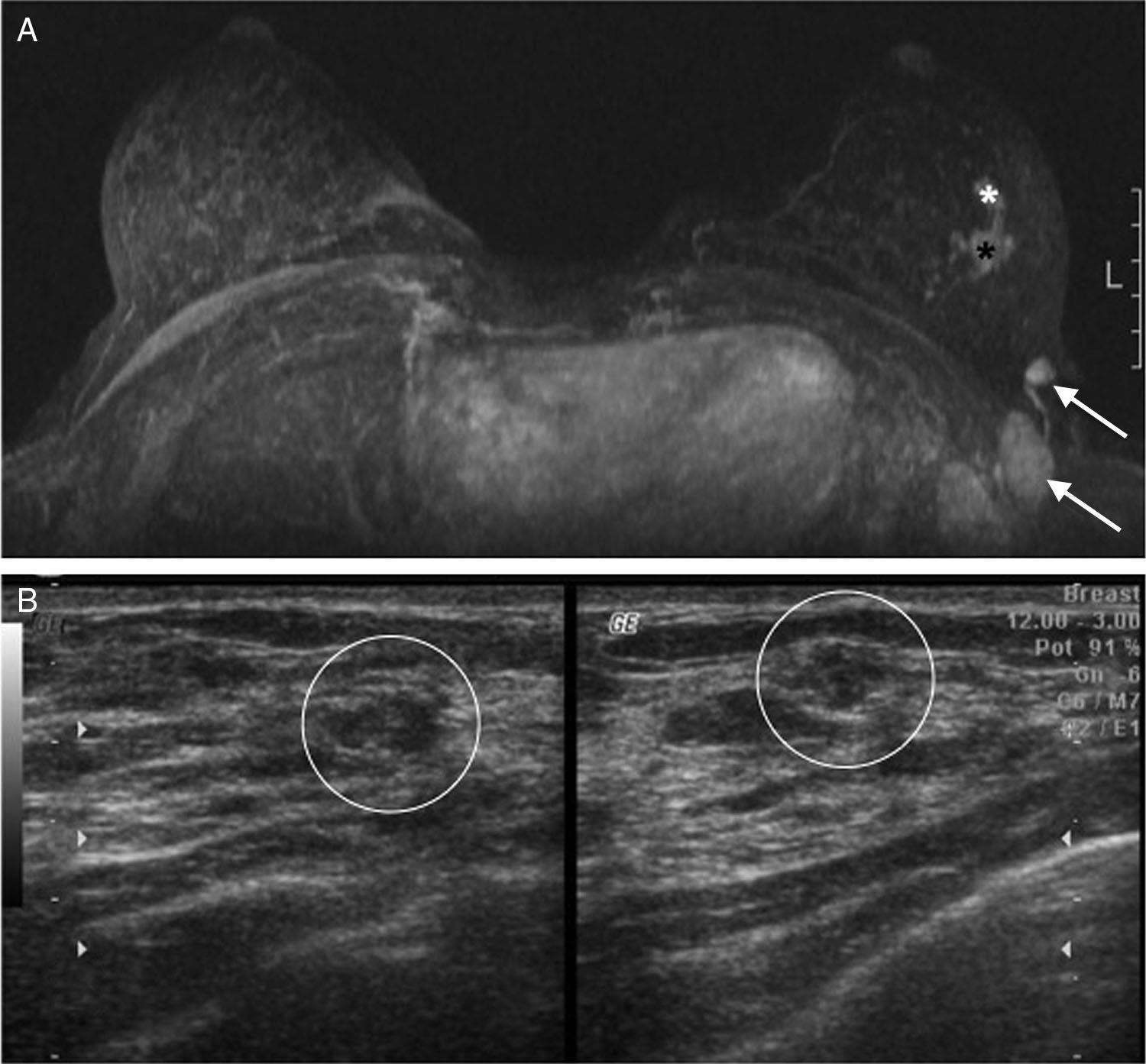
The only case in which the sample was not considered valid by the Dutch lab (though it was considered valid by our anatomopathologist) belonged to the largest tumor (5cm) in our series (Fig. 3). The possibility of having performed a biopsy on necrotic areas of the tumor could just be a hypothesis.
Case #6: 53-year-old patient who went to the doctor after finding one right “lump” several months old. Both in the ultrasound (A) and the MRIs (B) one 5cm mass (*) that seems to contain heterogeneous areas inside (arrow) maybe due to necrosis foci can be seen. Determination through MammaPrint®: not valid. We then proceeded to perform to analyze the surgical sample: high risk.
The case in which the collection had to be repeated was a 1cm-tumor (Fig. 4). However, in case #3 (Fig. 1), with a tumor size of 0.6cm, the sample was considered valid. Therefore and according to our experience we cannot claim that the percentage of valid samples has a direct relation with tumor size.
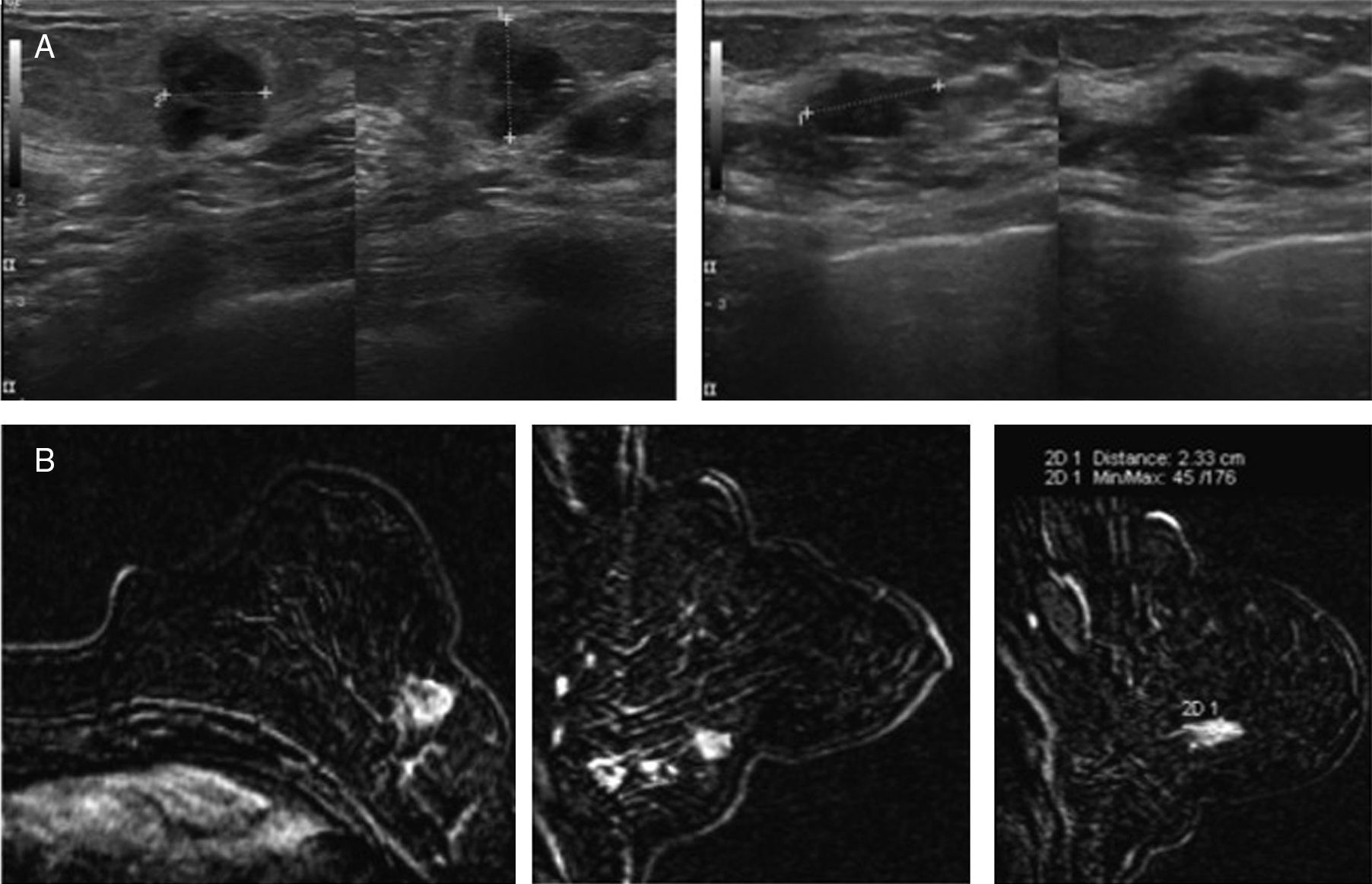
Case #1: 47-year-old patient who has been complaining of a left axillary lump for several months now with a normal mammogram and breast exam. The axillary ultrasound examination shows prominent adenopathies that were then submitted to biopsy with results of metastasis of probable primary breast tumor. One MRI (A) is performed that confirms the adenopathies (arrows) and shows 2 adjacent masses (*) of suspicious characteristics. The ultrasound-guided examination (B) shows subtle findings (circles) related to the MRI findings. One ultrasound-guided percutaneous biopsy of the largest focus (1cm) is performed confirming the presence of an infiltrating carcinoma. However according to the pathologist the sample did not contain enough material for MammaPrint® determination so the biopsy had to be repeated. Determination through MammaPrint®: high risk.
In all cases of valid samples, it has been possible to collect percentages of tumor cells equal to or greater than 60%, way above the limit required by the laboratory (30%) though we cannot say that the percentage achieved has a clear correlation with the size of the tumor. Thus 60% was obtained in tumors whose size ranged from 0.6 to 3.2cm, and 70% in tumors of 1.3–2.6cm in size.
The gauge of the needle used should be taken into consideration too. In the work by Osaki et al.,19 using ultrasound-guided VAB with 11 G needles, 3 periods are described among the total number of cases and in two of them (of 16 and 28 patients) we can see the percentages of valid samples of up to 87.5% and 85.7%, respectively that are discretely greater than ours. It would seem logical to admit that a larger percentage of tumor tissue could be obtained from samples collected with 11 G needles, especially if it is through VAB.
Another factor to take into consideration would be the number of samples collected, which in our experience has ranged from 3 to 4. The bibliographic data is ambiguous on this regard. Either there is no record of this information18,20 or there are references that talk about a very variable number of samples.19 In accordance with our results, we believe that 3 or 4 samples, with 12 G needles, could be enough considering that it is a procedure of initially diagnostic purposes.
Performing 3–4 passes with 12 G needles in ultrasound-guided VABs has been the routine of our group for some years now in view of lesions suspicious of malignancy, because it is customary to request immune-histochemical determinations, something feasible from the samples obtained with the aforementioned gauge. This is who the MammaPrint® determination did not change our routine protocol which by the way based on the results seems adequate if we wish to obtain the tumor genotype.
When it comes to the “element operator” in our study only one radiologist was involved just like in the periods in which the Osaki et al.19 group got the best results so this may probably be a factor as important or even more than the needle gauge or size of the tumor. In this sense we recommend to take samples in several areas of the tumor precluding necrotic areas (though these ones do not always show a clear ultrasound semiology) and avoid repeating the collection of samples in the same tumor region.
Yet despite the limitations of this study (retrospective and with few cases), and given the high proportion of valid samples (84.62%) we conclude that it is possible to obtain the genetic profile of breast cancer from the paraffin blocks of diagnostic PB, in almost all cases of malignant masses with ultrasound translation, with 3–4 samples obtained with 12 G needles if the operator is adequately experienced.
We believe that the results obtained make it possible to contemplate the possibility to add this usefulness to the diagnostic PB and also the results allow us to reinforce the role played by the radiologist in the therapeutic planning of breast cancer.
Ethical disclosuresProtection of human and animal subjectsThe authors declare that no experiments were performed on humans or animals for this investigation.
Confidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Author contributions- 1.
Manager of the integrity of the study: JALR, IZE and JAMA.
- 2.
Study idea: JALR.
- 3.
Study design: JALR.
- 4.
Data mining: JALR, IZE and JAMA.
- 5.
Statistical analysis: JALR.
- 6.
Reference search: JALR and JAMA.
- 7.
Writing: JALR, IZE and JAMA.
- 8.
Critical review of the manuscript: JALR, IZE and JAMA.
- 9.
Approval of final version: JALR, IZE and JAMA.
The authors declare no conflict of interests associated with this article whatsoever.
Please cite this article as: López Ruiz JA, Zabalza Estévez I, Mieza Arana JA. Cáncer de mama: determinación del perfil genético a partir de la biopsia percutánea ecoguiada diagnóstica. Radiología. 2016;58:214–220.