Mucor circinelloides is an opportunistic fungus capable of causing mucormycosis, a highly aggressive infection of quick spreading. Besides, it also has a high mortality rate due to late diagnosis and difficult treatment.
AimsIn this study we have identified the most immunoreactive proteins of the secretome and the total protein extract of M. circinelloides using sera from immunocompromised infected mice.
MethodsThe proteins of the secretome and the total extract were analyzed by two-dimensional electrophoresis and the most immunoreactive antigens were detected by Western Blot, facing the sera of immunocompromised infected mice to the proteins obtained in both extracts of M. circinelloides.
ResultsSeven antigens were detected in the secretome extract, and two in the total extract, all of them corresponding only to three proteins. The enzyme enolase was detected in both extracts, while triosephosphate isomerase was detected in the secretome, and heat shock protein HSS1 in the total extract.
ConclusionsIn this work the most immunoreactive antigens of the secretome and the total extract of M. circinelloides were identified. The identified proteins are well known fungal antigens and, therefore, these findings can be useful for future research into alternatives for the diagnosis and treatment of mucormycosis.
Mucor circinelloides es un hongo oportunista causante de la mucormicosis, una infección altamente agresiva y de rápida expansión. Además, también presenta una alta mortalidad debido al diagnóstico tardío y el difícil tratamiento.
ObjetivosEn este estudio se han identificado las proteínas más inmunorreactivas del secretoma y del extracto total de proteínas de M. circinelloides mediante el uso de sueros obtenidos de ratones inmunodeprimidos infectados.
MétodosLas proteínas del secretoma y del extracto total se analizaron mediante electroforesis bidimensional y se detectaron los antígenos más inmunorreactivos mediante Western Blot, enfrentando el suero de los ratones inmunodeprimidos infectados a las proteínas obtenidas en ambos extractos de M. circinelloides.
ResultadosSe identificaron 7 antígenos en el secretoma y 2 en el extracto total, todos ellos correspondientes a 3 proteínas. La enolasa se detectó en ambos extractos, mientras que la triosafosfato isomerasa se detectó en el secretoma, y la proteína de choque térmico HSS1 en el extracto total.
ConclusionesEn este trabajo se identificaron los antígenos más inmunorreactivos del secretoma y del extracto total de M. circinelloides. Todas las proteínas identificadas son antígenos fúngicos muy conocidos y, por ello, estos resultados pueden ser de gran utilidad en futuras investigaciones relacionadas con la mejora del diagnóstico y el tratamiento de la mucormicosis.
Mucor circinelloides is a saprophyte and pathogenic fungus which affects, above all, immunocompromised individuals, both human and animal. According to a recent review of cases of mucormycosis between January 2000 and January 2017, Mucor species are the second most common causal agent of mucormycosis, only outnumbered by fungi within the genus Rhizopus.9 In the last few years the incidence of mucormycosis has increased, the principal cause being the marked rise in debilitating diseases of the immune system, which includes pathologies such as bone marrow or solid organ transplantations, immunodeficiencies related to HIV, diabetes and primary or iatrogenic neutropenia.10 Recently, it has also caused an outbreak in burned patients.6 Mucormycosis can occur by inhalation of airborne conidia or inoculation on an open wound,18 it is highly aggressive and spreads quickly, invading the blood vessels, causing hemorrhages, thrombosis, heart attacks and tissue necrosis. In addition, it must be taken into account the fact that diagnosis is usually carried out at a late stage of infection, and that treatment strategies are not clear.21,22 Because of these reasons outlined above and the fact that it is an emerging pathology, more research into this disease is of paramount importance.7 Therefore, the aim of this work was to achieve the identification of the most immunogenic antigens of M. circinelloides. To do that, an innovative approach using immunosuppressed mice was employed to detect them so that only the most immunoreactive were selected.
Materials and methodsStrains and culture conditionsThe strain of M. circinelloides used in this study was CBS 125721, isolated from human maxillary tissue. It was cryopreserved at −80°C and cultured onto Potato Dextrose Agar (PDA) (Pronadisa, Madrid, Spain) at 28°C for 7 days before use. The conidiospores were obtained by washing the plates with sterile saline (0.9% NaCl), filtered through gauze and centrifuged. The concentration of conidiospores was adjusted as needed using a hemocytometer.
Animal modelsThe sera used for the detection of the most immunoreactive antigens were obtained from two groups of 5 mice each immunosupressed with cyclophosphamide: one was infected with fungal conidia, and the other one was injected with saline solution, according to a method previously described.13 Briefly, the mice were immunosuppressed 2 days prior the infection by intraperitoneal injection of 200mg/kg body weight of cyclophosphamide and once every 5 days thereafter, and inoculated intravenously with a density of 105 conidia or saline solution and sacrificed 20 days later. All experiments involving animals were in accordance with the ethical standards of Universitat Rovira i Virgili Animal Welfare and Ethics Comittee.
Obtention of total protein extract and secretome of M. circinelloidesFor the total extract and the secretome, 5×105conidia/ml and 106conidia/ml were grown, respectively, for 24h at 37°C and 120rpm in 150ml Potato Dextrose Broth (PDB). After culture, both conidia and hyphae morphologies were obtained. Then, the fungus was centrifuged (12,000×g, 5min, 4°C), and washed with PBS.
For the total extract the pellet was resuspended in PBS with 1% β-mercaptoethanol and 1% pharmalytes, then it was lysed using crystal beads (0.5mm diameter) at 30Hz for 20min using the Millmix 20 Bead-Beater (Thetnica, Eslovenia, Europe).
The secretome was obtained as described by da Silva et al.5 for Scedosporiumboydii, with a few modifications. After growing in PDB, the fungus was collected and cultured in PBS-2% glucose for 20h at 37°C and 120rpm. Finally, the obtained culture was centrifuged at 11,000×g for 20min and the supernatant was sterilized through 0.22μm filters (Merck Millipore, MAS, USA). To confirm the absence of cytoplasmic proteins in the secretome the activity of the intracellular enzyme lactate dehydrogenase (LDH) was measured, as previously described,23 and the cellular integrity of the fungus was observed under optical microscopy.
Protein detection by two-dimensional electrophoresisThe proteins were precipitated and two-dimensional electrophoresis (2-DE) was carried out following the protocol described in a previous work,15 using 18cm long Immobiline DryStrip gels (pH 3–10, GE Healthcare). The conditions of the isoelectric focusing were modified for the secretome: 12h rehydration, 150V for 300Vhr (Step and Hold, S&H), 500V for 2000Vhr (S&H), 1000V for 9000Vhr (gradient), 8000V for 20,000Vhr (gradient) and 8000V for 100,000Vhr (S&H). The second dimension was carried out in the PROTEAN II xi Cell system (Bio-Rad, CA, USA) at 45mA using 10% polyacrylamide gels. The 2-DE gels were stained with Coomassie Brilliant Blue to visualize proteins.
Antigen detectionProteins were electrotransferred to Hybond-P PVDF membranes, which were stained with Ponceau red to check the correct transfer of the proteins. Then, Western Blot (WB) was performed in accordance with the protocol described by Pellon et al.15 using the sera of infected mice, pooled and diluted 1/100 as sample. The experiments were carried out in triplicate and the most immunoreactive antigens were selected using ImageMaster 2D Platinum software (GE Healthcare).
Identification of immunoreactive proteinsThe identified spots were extracted from the gels for identification by mass spectrometry (LC–MS/MS) in the service of proteomics SGIker of the University of the Basque Country (UPV/EHU), as described by Buldain et al.2 Briefly, the extracted gel pieces were swollen in digestion buffer (50mM NH4HCO3, 12.5ng/μl trypsin [Roche, Basel, Switzerland]) and incubated at 37°C overnight. After that, the peptides were extracted, first with 25mM NH4HCO3 and acetonitrile (ACN), and secondly with 0.1% (v/v) trifluoroacetic acid and ACN. LC–MS/MS was carried out on a SYNAPT HDMS mass spectrometer (Waters, Milford, MA, USA) interfaced with a nanoAcquity UPLC System (Waters). The search for protein identification was made in the non-redundant database of the NCBI, restricted to fungi, using the online server MASCOT (Matrix Science Ltd., London, UK (http://www.matrixscience.com).
Bioinformatic analysis of M. circinelloides antigensIn order to study the relation between the identified antigens and their homologues in other organisms, the BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used. Then, the similarity values found between the proteins were compared.
ResultsInoculation of immunosuppressed mice with M. circinelloidesIn this study, mice immunosuppressed with cyclophosphamide were used to reduce the immune response and, in consequence, to avoid excessive immunoreactivity in WBs. Besides, we selected a strain of M. circinelloides previously classified as avirulent, which allowed us to collect the sera from all mice as the mortality rate in both infected and non-infected groups was zero. All the data of the infection process using this avirulent strain were described in a previous study.13
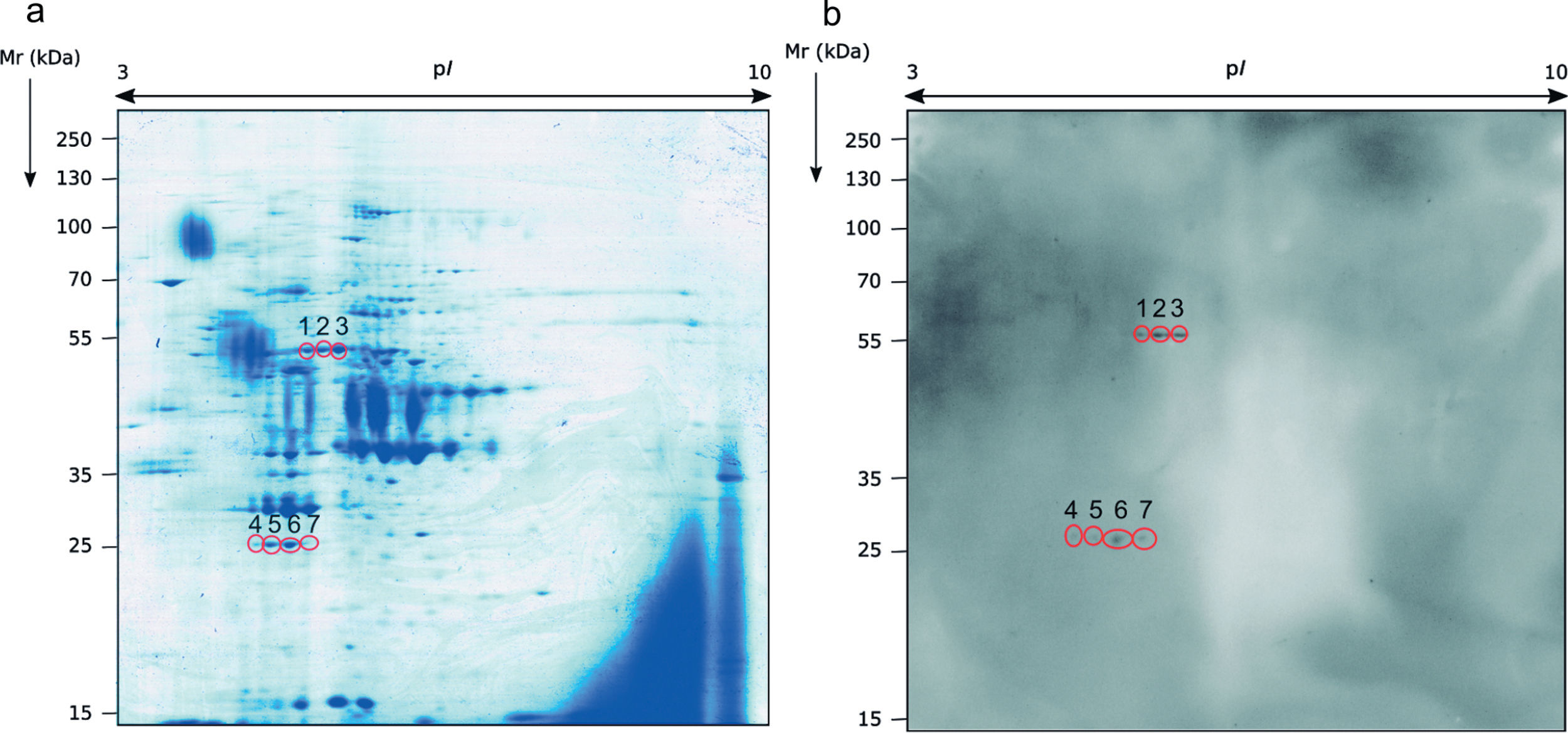
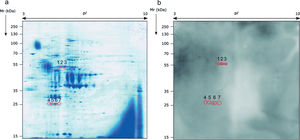
Identification of the immunoreactive antigens of the secretome recognized by sera of immunocompromised infected miceThe proteomics study of the secretome of M. circinelloides by 2-DE showed that secreted proteins were localized throughout the whole isoelectric point (pI) range used and with molecular weights (Mr) smaller than 130kDa. Specifically, the majority of the proteins contained in the secretome presented a pI between 4.5 and 7, and a Mr between 25 and 70kDa (Fig. 1a).
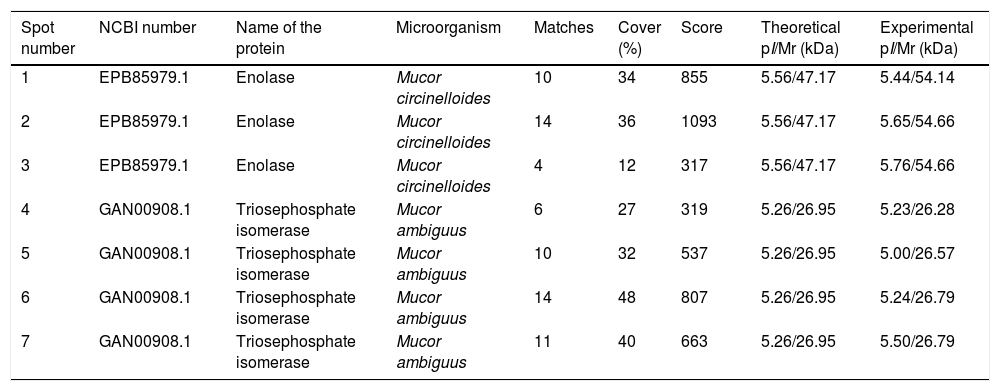
Sera of immunocompromised infected mice were used against the obtained extract; the immunoreactivity was analyzed and the seven most immunoreactive antigens were identified by mass spectrometry (Fig. 1). Three of these spots corresponded to enolase and four to triosephosphate isomerase, as shown in Table 1. They are almost certainly isoforms of the enolase and triosephosphate isomerase, respectively, as they have the same molecular weights and very similar isoelectric points, as it can be observed in Fig. 1a. The sera of mice injected with saline solution were also used at the same dilution and, as expected, no reactivity was observed (data not shown).
Analysis of the antigens identified in the secretome of Mucor circinelloides. Different parameters of the identifications are shown.
| Spot number | NCBI number | Name of the protein | Microorganism | Matches | Cover (%) | Score | Theoretical pI/Mr (kDa) | Experimental pI/Mr (kDa) |
|---|---|---|---|---|---|---|---|---|
| 1 | EPB85979.1 | Enolase | Mucor circinelloides | 10 | 34 | 855 | 5.56/47.17 | 5.44/54.14 |
| 2 | EPB85979.1 | Enolase | Mucor circinelloides | 14 | 36 | 1093 | 5.56/47.17 | 5.65/54.66 |
| 3 | EPB85979.1 | Enolase | Mucor circinelloides | 4 | 12 | 317 | 5.56/47.17 | 5.76/54.66 |
| 4 | GAN00908.1 | Triosephosphate isomerase | Mucor ambiguus | 6 | 27 | 319 | 5.26/26.95 | 5.23/26.28 |
| 5 | GAN00908.1 | Triosephosphate isomerase | Mucor ambiguus | 10 | 32 | 537 | 5.26/26.95 | 5.00/26.57 |
| 6 | GAN00908.1 | Triosephosphate isomerase | Mucor ambiguus | 14 | 48 | 807 | 5.26/26.95 | 5.24/26.79 |
| 7 | GAN00908.1 | Triosephosphate isomerase | Mucor ambiguus | 11 | 40 | 663 | 5.26/26.95 | 5.50/26.79 |
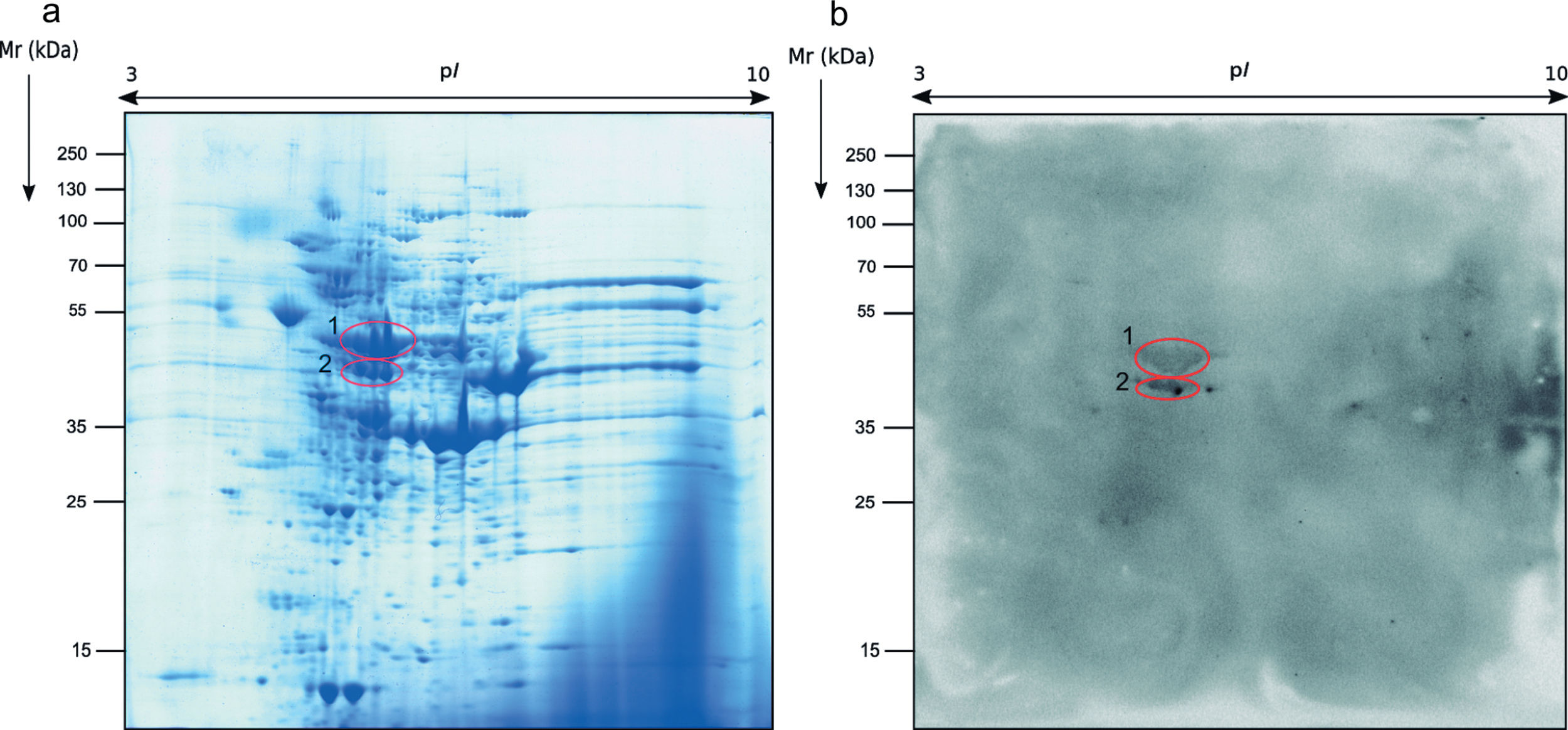
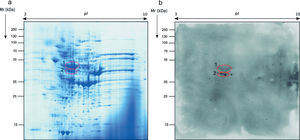
Regarding the study of the fungal cell proteome of the total extract by 2-DE, the proteins detected were also localized on the whole range of pI and Mr used. However, the protein distribution pattern observed was very different from the secretome and almost all proteins were present in the ranges of pI of 5–9 and Mr of 40–70kDa (Fig. 2a).
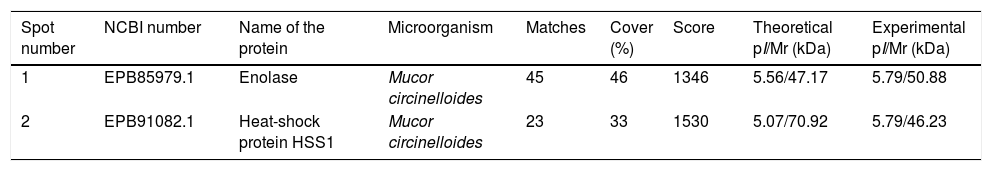
The immunome detected by two-dimensional WB showed a very few number of immunoreactive proteins recognized by sera from immunosuppressed mice and, therefore, only the two most immunoreactive spots were selected for their further identification (Fig. 2). These proteins were identified by mass spectrometry as enolase and heat shock protein HSS1 (Table 2). In the same way than for the secretome, the sera of mice injected with saline solution did not show any immunoreactivity (data not shown).
Analysis of the antigens identified in the total extract of Mucor circinelloides. Different parameters of the identifications are shown.
| Spot number | NCBI number | Name of the protein | Microorganism | Matches | Cover (%) | Score | Theoretical pI/Mr (kDa) | Experimental pI/Mr (kDa) |
|---|---|---|---|---|---|---|---|---|
| 1 | EPB85979.1 | Enolase | Mucor circinelloides | 45 | 46 | 1346 | 5.56/47.17 | 5.79/50.88 |
| 2 | EPB91082.1 | Heat-shock protein HSS1 | Mucor circinelloides | 23 | 33 | 1530 | 5.07/70.92 | 5.79/46.23 |
When comparing the sequence of the identified triosephosphate isomerase with the genome of M. circinelloides, an identity value of 99.48% was found. The Hsp70 identified in our work has similarity values of 78.58%, 76.43% and 66.39% with the proteins SSA1 and SSA2 found in Candida albicans, and the Hsp70 found in Rhizopus arrhizus, respectively. Finally, the sequence of the enolase of M. circinelloides was compared with the enolase of C. albicans, R. arrhizus and Homo sapiens, and similarity values of 69%, 81.88% and 71% were found, respectively.
DiscussionDue to the increase of immune system debilitating diseases in the last years, the prevalence of many infections has arisen. Mucormycosis is one of these infectious diseases, which can be caused by the fungus M. circinelloides. Mucormycosis is known for its high aggressiveness and quick spreading. Besides, due to the late diagnosis in the majority of the cases and the presence of high percentage of resistant isolates, this disease has high mortality rates.
Therefore, the aim of this work was to identify the most immunoreactive antigens of M. circinelloides, analyzing the immune response produced by sera obtained from immunocompromised infected mice. We used a cyclophosphamide-based immunosuppressive treatment to reduce the humoral immune response and, therefore, avoid the excess of signal which might mask and difficult the interpretation of the results. In this way, we proceeded to the identification of only the most immunogenic antigens by LC–MS/MS.
As a result, we identified in the secretome extract, from seven immunoreactive spots, two proteins, being them enolase and triosephosphate isomerase; in the total extract, from two spots, two proteins were identified, enolase and heat-shock protein HSS1. Among the proteins identified in the secretome extract, three isoforms of enolase and four of triosephosphate isomerase were detected. The spectrometric identifications showed that the proteins are probably the product of the same gene and, therefore, the little differences found among the pI values of these proteins might be due to slight modifications during the secretion process.
As enolase and triosephosphate isomerase, usually cytoplasmic proteins, were detected in the secretome, the mechanism of their secretion and function are not completely understood. The secretion process involved in M. circinelloides could be through extracellular vesicles, as seen with triosephosphate isomerase of the dimorphic fungus Paracoccidioides brasiliensis.12 Besides, taking into account the plasminogen binding activities of these two proteins, their presence in the external medium could be related to pathogen dissemination.4
Although triosephosphate isomerase was detected in the secretome extract of M. circinelloides, the identification by LC–MS/MS of the selected spot resulted in the fungus of the same genus Mucor ambiguus, because the used database determined this protein as the one found in this microorganism. When comparing this protein sequence with the genome of M. circinelloides, an identity value of 99.48% was found, indicating the high degree of similarity between the sequences of triosephosphate isomerase in both species. This protein was also identified in the fungi C. albicans19 and P. brasiliensis, and it is considered an important antigen, capable of binding the laminin and fibronectin of the extracellular matrix.17 Moreover, it can be found in the cell wall, but also in the secreted extracellular vesicles.12
Regarding HSS1, it belongs to the same heat shock protein family as the Hsp70,1 which in fungal species is overexpressed during infection to prevent the denaturalization of proteins14 and has been described as immunoreactive against serum IgGs15 and mucosal IgAs.16 In fact, HSS1 could be considered an orthologue of the Hsp70 proteins, SSA1 and SSA2, found in the ascomycetous fungi C. albicans, and the Hsp70 of R. arrhizus, as it shares with those proteins similarity values higher than 65%. Although there is a high percentage of homology, there are some specific regions when compared with the sequences of SSA1 of C. albicans and Hsp70 of R. arrhizus. This fact could be of great interest for diagnosis. Heat-shock proteins have been reported as antigens and virulence factors in the cell surface15 and the secretome of important pathogenic fungi, such as Lomentospora prolificans.3 In this sense, the location of the protein could ease the access of the immune cells, contributing to the high immunogenicity presented. Besides, it has been proved that the Hsp70 of Mycobacterium tuberculosis is capable of inducing the immune response, stimulating human monocytes to produce chemokines and cytokines.24 Given the major functions that Hsp70 has on infective processes and the homology between this protein and the one found in our study, it could be hypothesized that the protein HSS1 could have a similar role in the infections caused by M. circinelloides.
Enolase deserves a special mention as it has been identified in both analyzed protein extracts. Enolase has been also identified in the secretome of the mycelia and yeast cells of the dimorphic fungus Paracoccidioides lutzii.25 This metabolic enzyme was also previously identified by our research group as an antigen recognized by salivary IgA in L. prolificans,2 and by serum IgGs in C. albicans.8 Enolase was also associated with the cell wall in L. prolificans15 and C. albicans,14 where it performs transglutaminase activity, indicating the possible role of this enzyme in infection processes.20 Enolase, therefore, seems to be an important antigen for many of the most common pathogenic fungi, which makes it a reasonable target in panfungal diagnosis or a key to design new treatments or vaccines. In fact, the use of this enzyme for therapy was tested as a vaccine against C. albicans, lowering both the fungal burden and the amount of tissue damage.11
In order to determine the possible use of this protein in diagnosis or treatment strategies, its sequence was compared with the sequence in C. albicans, R. arrhizus and H. sapiens, and the similarity values found make probable a cross-reactivity with other fungi, as it happens between enolase of L. prolificans and Scedosporium apiospermum, S. boydii and Aspergillus fumigatus.14 Hence, although it could not be used for a specific diagnosis or therapeutic target, it could be used as a panfungal antigen.
In conclusion, in this work the most immunoreactive antigens of the secretome and the total extract of M. circinelloides were identified. The proteins identified were the HSS1 protein, along with enolase and triosephosphate isomerase, which are well-known fungal antigens. These proteins might be useful in the future for the development of a vaccine, antifungal treatments and/or for diagnosis, allowing the rapid detection and treatment of the disease and, therefore, lowering the unacceptable mortality rates.
FundingThis study was funded by the University of the Basque Country (UPV/EHU) [grant number PPG17/41] and by the Basque Government [grant number IT1362-19]. MA and LMS have received a Grant from the Basque Government and LAF from the UPV/EHU.
Authors contributionsAll authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by MA, LMS and ARG. The first draft of the manuscript was written by MA and all authors commented on previous versions of the manuscript. All authors read and approved the final version.
Conflict of interestThe authors declare that they have no conflict of interest.
We thank the member of the Chartered of Linguists, No. 022913 for improving the English in the manuscript.