In this research, we evaluated the association of genetic variants rs1800629 in TNF and rs2228145 in IL6R with production of neutralising antibodies against SARS-CoV-2 and the frequency of adverse effects following immunisation (AEFIs) in adult population from Western of Mexico that received AZD1222 vaccination.
Methods117 adults were evaluated [33 years (23–40), 65% women]. The self-reported frequency of AEFIs was recorded and the percentage of post-vaccination neutralising antibodies was quantified. The identification of rs1800629 variant in TNF and rs2228145 in IL6R was performed by qPCR.
ResultsThe genotype frequencies of rs1800629 variant were: GG (86%) and GA (14%), and for rs2228145 variant were: AA (20%), CA (48%), and CC (32%). The percentage of post-vaccination antibodies was similar between men and women (median, 97.24%). An association was found between the frequency of AEFIs with the sex; being adynamia (P=.0243), chills (P=.0085), arthralgia (P=.0227), and pain in application area (P=.0096) more frequent in women. The GA genotype of rs1800629 variant showed an association with fever (P=.0131) and arthralgia (P=.0058) post-vaccination, but no relationship was found with the production of neutralising antibodies after AZD1222 vaccination. The rs2228145 variant was not associated with the production of antibodies nor with the AEFIs reported.
ConclusionThe genetic variants rs1800629 in TNF and rs222815 in IL6R are not associated with the production of neutralising antibodies against SARS-CoV-2 after receiving AZD1222 scheme in population from western of Mexico; however, the results suggest that rs1800629 variant increases the frequency of post-vaccination events, particularly fever and arthralgia.
En esta investigación se evaluó la asociación de las variantes genéticas rs1800629 en TNF y rs2228145 en IL6R con la producción de anticuerpos neutralizantes contra SARS-CoV-2 y la frecuencia de eventos supuestatemente atribuibles a la vacunación o inmunización (ESAVIs) en población adulta del occidente de México con esquema de vacunación AZD1222.
MétodosSe evaluaron 117 adultos [33 años (23–40), 65% mujeres]. Se registró la frecuencia autoreportada de ESAVIs y se cuantificó el porcentaje de anticuerpos neutralizantes post-vacunación. La identificación de las variantes genéticas rs1800629 y rs2228145 se realizó mediante qPCR.
ResultadosLa frecuencia de los genotipos de la variante rs1800629 en TNF fue: GG (86%) y GA (14%) y de la variante rs2228145 en IL6R: AA (20%), CA (48%) y CC (32%). El porcentaje de anticuerpos post-vacunación fue similar entre hombres y mujeres (mediana 97.24%). La frecuencia de los ESAVIs adinamia (p = 0.0243), escalofríos (p = 0.0085), artralgia (p = 0.0227) y dolor en zona de aplicación (p = 0.0096) fueron más frecuentes en mujeres. El genotipo GA de rs1800629 se asoció con fiebre (p = 0.0131) y artralgia (p = 0.0058) post-vacunación, pero no con la producción de anticuerpos neutralizantes. La variante rs2228145 no se asoció con la producción de anticuerpos ni con la frecuencia de ESAVIs.
ConclusiónLas variantes rs1800629 en TNF y rs222815 en IL6R no se asocian con la producción de anticuerpos neutralizantes contra SARS-CoV-2 después de recibir el esquema AZD1222 en población del occidente de México, sin embargo, los resultados sugieren que la variante rs1800629 incrementa la frecuencia de los ESAVIs fiebre y artralgia.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic that caused COVID-19 and its rapid spread worldwide fuelled the development and administration of vaccines in a short period of time to control the spread of the virus and reduce COVID-19's severity and mortality.1 Mexico reported one of the highest case fatality rates worldwide (11%–13%) and despite there being a number of factors associated with increased mortality,2 host genetics has been implicated in the susceptibility to and severity of infection and in the immune response triggered by vaccines, as well as in post-vaccination effects.3
In western Mexico, the AZD1222 vaccine (AstraZeneca, Oxford University) has been one of the most widely used; it is based on a non-replicative recombinant adenovirus-like viral vector containing encoded information to produce the SARS-CoV-2 spike (S) protein, stimulating humoral and cellular immune responses.4 The S protein is an important antigenic site to which the neutralising antibodies produced by the host bind in order to prevent the virus from entering5 and is therefore crucial in the design of vaccines such as AZD1222. The Events Supposedly Attributable to Vaccination or Immunisation (ESAVI) denote an unfavourable and untoward clinical event following vaccination, not necessarily causally related to the immunisation process or the vaccine itself.6 The most common ESAVI associated with the AZD1222 vaccine are pain at the site of injection, headache, muscle ache, fatigue, joint pain, and fever.4
On the other hand, tumour necrosis factor alpha (TNF-α) is a proinflammatory cytokine and a key regulator of the innate, adaptive immune system; it is involved in the recruitment of lymphocytes to areas of infection, activates antigen-presenting cells, and can activate B cells through the T cell-independent pathway, enabling their proliferation and differentiation into antibody-producing plasma cells.7 The encoding gene is TNF and it is highly polymorphic. The rs1800629 variant is located in the promoter region of the TNF gene and has been proposed as having an effect on gene expression, inasmuch as TNF-α expression is greater in individuals with GA and AA genotypes than in carriers of the GG genotype.8 For its part, the IL-6 cytokine is an important inducer of the response in the acute phase that regulates genes involved in cell differentiation, survival, apoptosis, and proliferation; for signalling, it must interact with the IL-6 receptor (IL-6R), which can be present in 2 forms: transmembrane and soluble.9 The gene encoding for this receptor is IL6R and is highly polymorphic.10 The genetic variant rs2228145 is located in exon 9 of IL6R and results in an amino acid substitution at the protein level (aspartic acid [Asp] for alanine [Ala]) at position 358 within the extracellular domain of IL-6R11.11 It has been suggested that this polymorphism leads to greater IL-6R shedding from the cell surface, thereby causing an alteration of the classical IL-6 pathway12 and allele C (358 Ala) has been associated with increased concentrations of circulating sIL-6r.13
The immune response generated after vaccination may be affected by the presence of variants in genes of the immune system, as has been previously reported for measles, rubella, and hepatitis B vaccines, among others.3 In this regard, the presence of the AA polymorphic genotype of rs1800629 in TNF was previously associated with low antibody production from the hepatitis B vaccine.14 Another study revealed a low rate of seroconversion in carriers of this variant when they received the Japanese encephalitis vaccine, although it was not significant.15 Meanwhile, the C allele of the rs2228145 polymorphism in IL6R has been linked to diseases such as amyotrophic lateral sclerosis,16 as well as the development of early-onset Alzheimer's disease12 and the development of type 1 diabetes in adulthood.17
There are currently no studies that associate the rs1800629 and rs228145 variants in TNF and IL6R with the production of neutralising anti-SARS-CoV-2 antibodies after receiving the AZD1222 vaccination regimen. Moreover, the response of immunised individuals is heterogeneous and there is insufficient data to determine antibody production after receiving this regimen. Therefore, the aim of this research was study of the adult population of western Mexico that was vaccinated with the AZD1222 scheme, with the goal of determining whether both variants in immune response genes constitute a genetic factor associated with the production of neutralising antibodies generated by the AZD1222 vaccine and with the frequency of ESAVI. Research into the immunogenetic factors associated with the post-vaccination response is worthwhile, as it may explain the heterogeneity of this response in the study population.
Material and methodStudy design and locationThe present study consisted of a cross-sectional-analytical design. Participants attended the Laboratorio de Biomedicina y Biotecnología para la Salud en el Centro Universitario del Sur de la Universidad de Guadalajara (Laboratory of Biomedicine and Biotechnology for Health at the Southern University Centre of the University of Guadalajara) (October 2021–February 2022), where a blood sample was obtained in order to separate serum and extract genomic DNA (gDNA). During the same visit, a sociodemographic data survey was administered, specifically the ESAVI frequency questionnaire, and an anthropometric evaluation of the participants were carried out.
ParticipantsParticipants were recruited by invitation. Sample size calculation was performed with the OpenEpi programme (v. 3), using the genotypic frequencies reported by SNPedia in the population of Mexican ancestry (residing in Los Angeles, USA) for the rs1800629 variant. A sample of 116 was obtained, providing a statistical a power of 80% and a confidence level of 90%. A total of 117 participants were included, 38% male (n=45) and 62% female (n=72), with ages ranging from 18 to 60 years. Inclusion criteria consisted of having a 2-dose AZD1222 vaccination schedule, being at least 15 days after the last dose, and being of Mexican mixed race, with ancestry of at least 3 previous generations from Western Mexico. Exclusion criteria comprised: having a different SARS-CoV-2 vaccination schedule(s), being pregnant, receiving treatment with immunosuppressants, presenting symptoms, or confirmed SARS-CoV-2 infection or other infectious diseases at the time of sampling.
Body composition and anthropometricsAnthropometric assessment of the participants was performed following the guidelines laid down by the International Society for the Advancement of Kinanthropometry (ISAK). Weight (kg) and body fat (%) were measured with a body fat tracker and bioimpedance scale (BF-679W, Tanita, Japan), height (cm) with a portable stadiometer (213 l, SECA, USA), while waist and hip circumference (cm) were measured with an anthropometric tape (Lufkin, USA).
Quantification of neutralising antibodies against the SARS-CoV-2 S-proteinBlood samples were collected from participants by venipuncture into serum tubes. The sera were separated according to standardised procedures and stored at −60 °C. Quantification of the percentage of neutralising antibodies against SARS-CoV-2 was performed using the cPass SARS-CoV-2 Neutralisation Antibody Detection Kit (GenScript, China) based on a blocking enzyme-linked immunosorbent assay (ELISA). The protocol was carried out in accordance with the manufacturer's instructions. The absorbance reading was at 450 nm and neutralisation percentages were calculated using the formula provided by the manufacturer: neutralisation percentage=1-(Optical density [OD] value of the sample/OD value of the negative control)×100.
Genomic DNA extractionA blood sample was extracted by venipuncture, from which gDNA was extracted using the QIAamp DNA Blood Mini Kit (QIAGEN, Germany) following the methodology specified by the manufacturer. The quantity and quality of the gDNA obtained was verified by measuring the optical density at 260 and 280 nm in a spectrophotometer (Multiskan, Thermo Fischer Scientific, USA).
Identification of the genotypes for the rs1800629 and rs2228145 variantsThe rs1800629 and rs2228145 variants were identified by real-time polymerase chain reaction (qPCR) amplification using Taqman probes (C_7514879_10 and C_16170664_10, Applied Biosystems, USA). Reagents were prepared as per the manufacturer's recommendations as follows: 5 μl of TaqMan Master Mix; 0.5 μl TaqMan SNP Genotyping Assay; ∼50 ng gDNA; ∼3.5 μl of sterile water, to make up a total volume of 10 μl per reaction. The qPCR was carried out on the QuantStudio5 thermal cycler (Applied Biosystems, USA), with the following conditions: polymerase activation at 95 °C for 10 min, denaturation at 95 °C for 15 s, and alignment and extension at 60 °C for 1 min; all of the above times 40 cycles. DNAase-free water and DNA samples with known genotypes were used as negative control and positive controls, respectively. Genotype identification was undertaken using QuantStudio Design & Analysis Software v1.5.2 (Applied Biosystems, USA).
Statistical analysisData were analysed and plotted using the GraphPad Prism Ve.8.0.1 software (GraphPad Software LLC). Data distribution was checked by means of the Kolmogorov–Smirnov normality test and the D'Agostino normality test. Comparative analyses of antibody levels were performed based on sex or genotype; normally distributed continuous variables were compared by parametric tests (Student's t-test or ANOVA), while those that exhibited non-normal distribution were compared by non-parametric tests (Mann–Whitney U or Kruskal–Wallis). Categorical variables were expressed as percentages and frequencies were analysed by chi-squared test or Fisher's exact test, as appropriate. Significance was set at a level of P<.05.
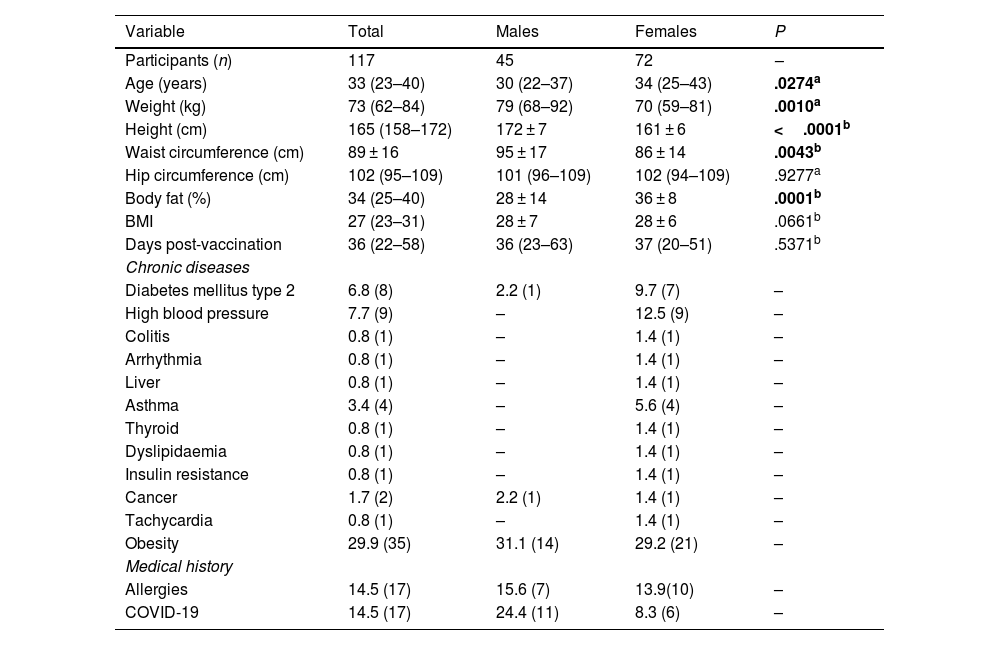
ResultsParticipants' sociodemographic and anthropometric characteristics and medical historyThe participants' sociodemographic and anthropometric characteristics and data from the clinical history are presented in Table 1. A total of 117 individuals were evaluated at a median of 36 (22–58) days post-vaccination, which were separated on the basis of subjects' sex. Significant differences were noted with respect to age (P=.0274) and body fat percentage (P=.0001), both of which were higher among the females. Weight (P=.0010), height (P<.0001), and waist circumference (P=.0043) were greater in the male group.
Participants' demographic and anthropometric data and clinical history data with the AZD1222 vaccination regimen.
| Variable | Total | Males | Females | P |
|---|---|---|---|---|
| Participants (n) | 117 | 45 | 72 | – |
| Age (years) | 33 (23–40) | 30 (22–37) | 34 (25–43) | .0274a |
| Weight (kg) | 73 (62–84) | 79 (68–92) | 70 (59–81) | .0010a |
| Height (cm) | 165 (158–172) | 172 ± 7 | 161 ± 6 | <.0001b |
| Waist circumference (cm) | 89 ± 16 | 95 ± 17 | 86 ± 14 | .0043b |
| Hip circumference (cm) | 102 (95–109) | 101 (96–109) | 102 (94–109) | .9277a |
| Body fat (%) | 34 (25–40) | 28 ± 14 | 36 ± 8 | .0001b |
| BMI | 27 (23–31) | 28 ± 7 | 28 ± 6 | .0661b |
| Days post-vaccination | 36 (22–58) | 36 (23–63) | 37 (20–51) | .5371b |
| Chronic diseases | ||||
| Diabetes mellitus type 2 | 6.8 (8) | 2.2 (1) | 9.7 (7) | – |
| High blood pressure | 7.7 (9) | – | 12.5 (9) | – |
| Colitis | 0.8 (1) | – | 1.4 (1) | – |
| Arrhythmia | 0.8 (1) | – | 1.4 (1) | – |
| Liver | 0.8 (1) | – | 1.4 (1) | – |
| Asthma | 3.4 (4) | – | 5.6 (4) | – |
| Thyroid | 0.8 (1) | – | 1.4 (1) | – |
| Dyslipidaemia | 0.8 (1) | – | 1.4 (1) | – |
| Insulin resistance | 0.8 (1) | – | 1.4 (1) | – |
| Cancer | 1.7 (2) | 2.2 (1) | 1.4 (1) | – |
| Tachycardia | 0.8 (1) | – | 1.4 (1) | – |
| Obesity | 29.9 (35) | 31.1 (14) | 29.2 (21) | – |
| Medical history | ||||
| Allergies | 14.5 (17) | 15.6 (7) | 13.9(10) | – |
| COVID-19 | 14.5 (17) | 24.4 (11) | 8.3 (6) | – |
BMI: body mass index. The data are presented as the mean±standard deviation or median (P25%–P75%), according to their distribution. Significant values appear in bold. The test was used depending on data normality.
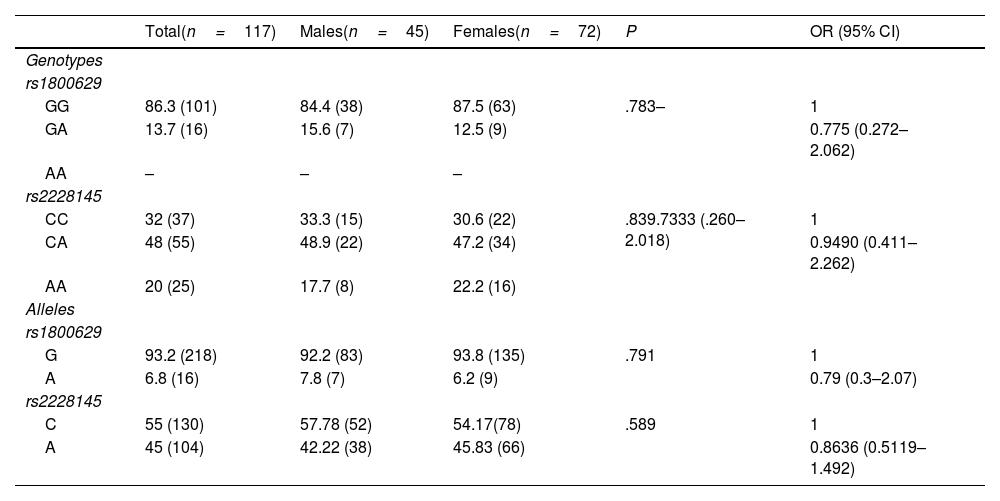
Two of the three genotypes of the rs1800629 variant were identified: the homozygous wild-type GG in 86.3% of the sample (n=101) and the heterozygous polymorphic GA in 13.7% (n=16); there were no cases of the homozygous polymorphic AA in the sample. The genotypic frequencies of this variant were found to be in Hardy–Weinberg equilibrium (P=.728). For the rs2228145 variant, all three genotypes were detected: the AA homozygote in 20% (n=25), the CA polymorphic heterozygote in 48% (n=55), and the polymorphic homozygote in 32% (n=37). It was determined that the genotypic frequencies of this variant were in Hardy–Weinberg equilibrium (P=.9243); it was therefore inferred that in the study population, there was a random selection, that there are no high mutation rates, and there was no migration or other phenomena that might cause its deviation. Genotypic and allelic frequencies of both variants are displayed in detail in Table 2. No significant differences were discovered when comparing genotypic and allelic frequencies on the basis of sex.
Genotypic and allelic frequencies by study group.
| Total(n=117) | Males(n=45) | Females(n=72) | P | OR (95% CI) | |
|---|---|---|---|---|---|
| Genotypes | |||||
| rs1800629 | |||||
| GG | 86.3 (101) | 84.4 (38) | 87.5 (63) | .783– | 1 |
| GA | 13.7 (16) | 15.6 (7) | 12.5 (9) | 0.775 (0.272–2.062) | |
| AA | – | – | – | ||
| rs2228145 | |||||
| CC | 32 (37) | 33.3 (15) | 30.6 (22) | .839.7333 (.260–2.018) | 1 |
| CA | 48 (55) | 48.9 (22) | 47.2 (34) | 0.9490 (0.411–2.262) | |
| AA | 20 (25) | 17.7 (8) | 22.2 (16) | ||
| Alleles | |||||
| rs1800629 | |||||
| G | 93.2 (218) | 92.2 (83) | 93.8 (135) | .791 | 1 |
| A | 6.8 (16) | 7.8 (7) | 6.2 (9) | 0.79 (0.3–2.07) | |
| rs2228145 | |||||
| C | 55 (130) | 57.78 (52) | 54.17(78) | .589 | 1 |
| A | 45 (104) | 42.22 (38) | 45.83 (66) | 0.8636 (0.5119–1.492) | |
The data are presented as percentages and frequency % (n). Chi-squared or Fisher's exact test as applicable. CI: confidence interval; OR: odds ratio.
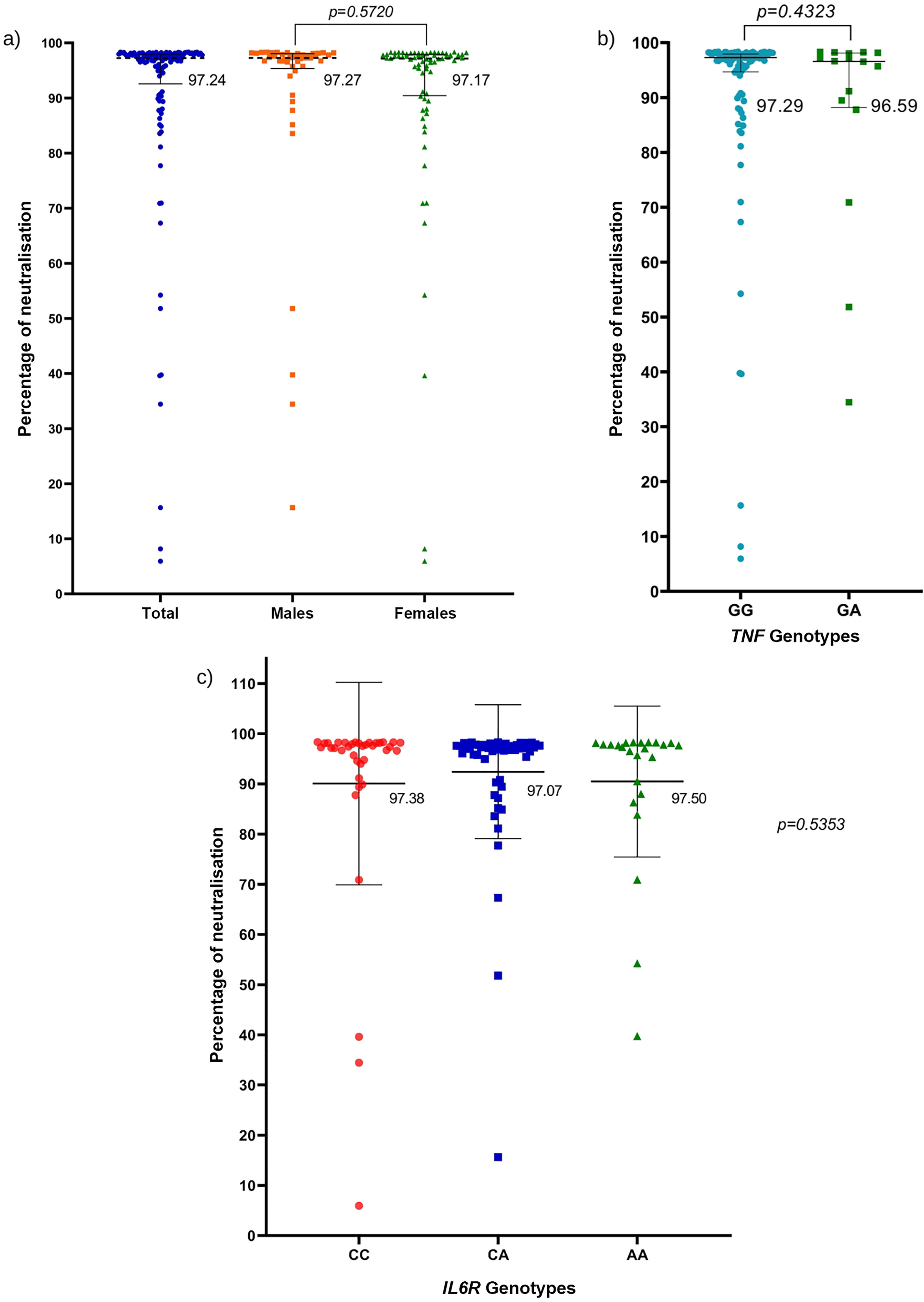
The percentages of neutralising antibodies generated against the SARS-CoV-2 S protein in the population with the AZD1222 regimen based on sex are illustrated in Fig. 1a. No statistical differences were detected with regard to the percentages of neutralisation between study groups.
a) Percentage of neutralisation against the SARS-CoV-2 spike protein by sex; Mann–Whitney U test. b) Percentages of neutralisation against the SARS-CoV-2 S protein with respect to the genotypes of the rs1800629 variant; Mann–Whitney U test. c) percentages of neutralisation against the SARS-CoV-2 S protein with respect to the genotypes of the rs2228145 variant; Kruskal–Wallis test. The horizontal lines represent the median and interquartile range (P25%–P75%).
The percentages of neutralisation against the SARS-CoV-2 S protein were analysed according to the genotypes of the rs1800629 variant; they were very similar to each other, and no significant differences were found between the TNF genotypes (P=.4323) (Fig. 1b). Bearing in mind that most of the participants had very high percentages of neutralising antibodies post-AZD1222 vaccination (more than 90% neutralisation), a sub-analysis by genotype was performed on the TNF gene, including only those participants with antibody percentages below 90% neutralisation. Nevertheless, no significant relationship was observed between neutralisation percentages of less than 90% with genotypes of the rs1800629 variant (data not shown).
Analysis of neutralisation percentages against SARS-CoV-2 protein S was performed based on the rs2228145 variant genotypes; a similar median was obtained for all 3 genotypes. There were no significant differences based on IL6R genotypes (Fig. 1c).
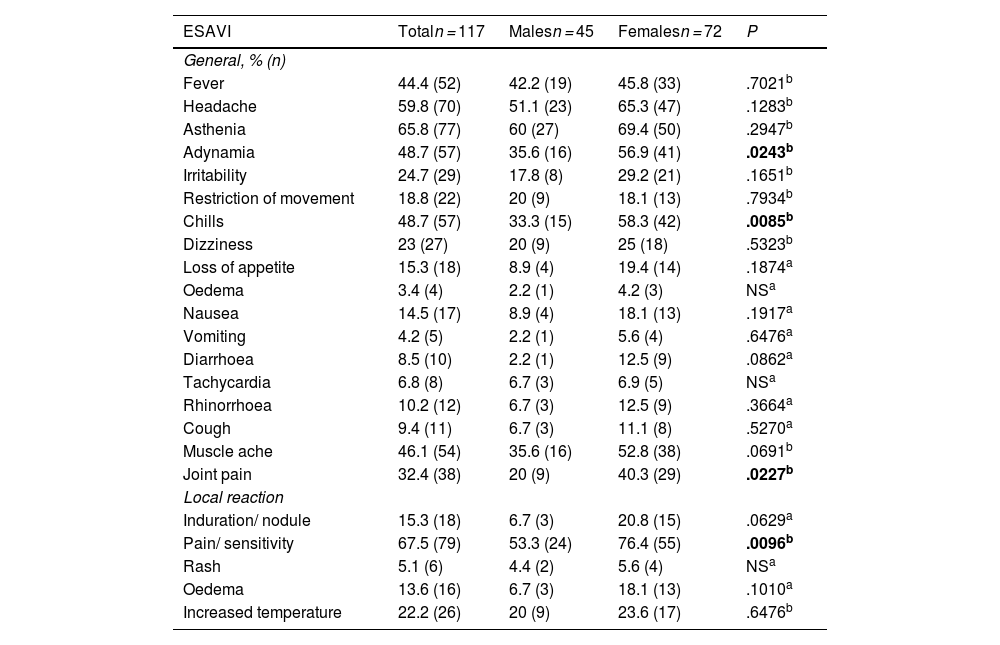
Frequency of events supposedly attributable to vaccination and immunisation: Association with sex and genotypes of the rs1800629 and rs2228145 variantsThe leading ESAVI reported by the participants are listed in Table 3. An association was found between sex and adynamia (P=.0243), chills (P=.0085), joint pain (P=.0227), and pain at the site of injection (P=.0096); all were more common among females. No association was detected between participants' sex and the remaining ESAVI.
Leading events supposedly attributable to vaccination or immunisation following AZD1222 vaccination by study group by sex.
| ESAVI | Totaln = 117 | Malesn = 45 | Femalesn = 72 | P |
|---|---|---|---|---|
| General, % (n) | ||||
| Fever | 44.4 (52) | 42.2 (19) | 45.8 (33) | .7021b |
| Headache | 59.8 (70) | 51.1 (23) | 65.3 (47) | .1283b |
| Asthenia | 65.8 (77) | 60 (27) | 69.4 (50) | .2947b |
| Adynamia | 48.7 (57) | 35.6 (16) | 56.9 (41) | .0243b |
| Irritability | 24.7 (29) | 17.8 (8) | 29.2 (21) | .1651b |
| Restriction of movement | 18.8 (22) | 20 (9) | 18.1 (13) | .7934b |
| Chills | 48.7 (57) | 33.3 (15) | 58.3 (42) | .0085b |
| Dizziness | 23 (27) | 20 (9) | 25 (18) | .5323b |
| Loss of appetite | 15.3 (18) | 8.9 (4) | 19.4 (14) | .1874a |
| Oedema | 3.4 (4) | 2.2 (1) | 4.2 (3) | NSa |
| Nausea | 14.5 (17) | 8.9 (4) | 18.1 (13) | .1917a |
| Vomiting | 4.2 (5) | 2.2 (1) | 5.6 (4) | .6476a |
| Diarrhoea | 8.5 (10) | 2.2 (1) | 12.5 (9) | .0862a |
| Tachycardia | 6.8 (8) | 6.7 (3) | 6.9 (5) | NSa |
| Rhinorrhoea | 10.2 (12) | 6.7 (3) | 12.5 (9) | .3664a |
| Cough | 9.4 (11) | 6.7 (3) | 11.1 (8) | .5270a |
| Muscle ache | 46.1 (54) | 35.6 (16) | 52.8 (38) | .0691b |
| Joint pain | 32.4 (38) | 20 (9) | 40.3 (29) | .0227b |
| Local reaction | ||||
| Induration/ nodule | 15.3 (18) | 6.7 (3) | 20.8 (15) | .0629a |
| Pain/ sensitivity | 67.5 (79) | 53.3 (24) | 76.4 (55) | .0096b |
| Rash | 5.1 (6) | 4.4 (2) | 5.6 (4) | NSa |
| Oedema | 13.6 (16) | 6.7 (3) | 18.1 (13) | .1010a |
| Increased temperature | 22.2 (26) | 20 (9) | 23.6 (17) | .6476b |
ESAVI: Events supposedly attributable to vaccination or immunisation. The data are presented as percentage and absolute frequency % (n). The significant values (P<.05) are displayed in bold.
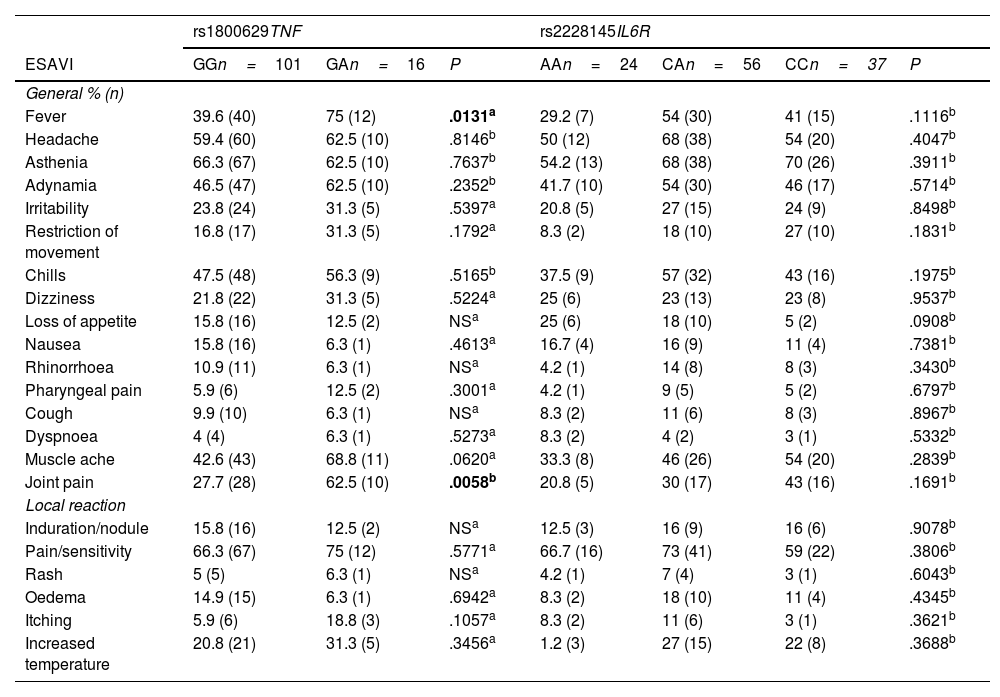
The most common ESAVI based on the genotypes of the rs1800629 variant of the TNF gene and the rs2228145 variant of the IL6R gene reported by the participants can be found in Table 4. An association was noted between the genotypes of the rs1800629 variant with the ESAVI fever (P=.0131) and joint pain (P=.0058), both of which were more frequent among participants in the group with the GA genotype compared to the GG genotype. No association was detected between the TNF rs1800629 variant and the remaining ESAVI. As regards the genotypes of the rs2228145 variant in IL6R, no association was identified with the ESAVI reported by the participants.
Leading events supposedly attributable to vaccination or immunisation following AZD1222 vaccination with respect to the genotypes of the rs1800629 and rs2228145 variants.
| rs1800629TNF | rs2228145IL6R | ||||||
|---|---|---|---|---|---|---|---|
| ESAVI | GGn=101 | GAn=16 | P | AAn=24 | CAn=56 | CCn=37 | P |
| General % (n) | |||||||
| Fever | 39.6 (40) | 75 (12) | .0131a | 29.2 (7) | 54 (30) | 41 (15) | .1116b |
| Headache | 59.4 (60) | 62.5 (10) | .8146b | 50 (12) | 68 (38) | 54 (20) | .4047b |
| Asthenia | 66.3 (67) | 62.5 (10) | .7637b | 54.2 (13) | 68 (38) | 70 (26) | .3911b |
| Adynamia | 46.5 (47) | 62.5 (10) | .2352b | 41.7 (10) | 54 (30) | 46 (17) | .5714b |
| Irritability | 23.8 (24) | 31.3 (5) | .5397a | 20.8 (5) | 27 (15) | 24 (9) | .8498b |
| Restriction of movement | 16.8 (17) | 31.3 (5) | .1792a | 8.3 (2) | 18 (10) | 27 (10) | .1831b |
| Chills | 47.5 (48) | 56.3 (9) | .5165b | 37.5 (9) | 57 (32) | 43 (16) | .1975b |
| Dizziness | 21.8 (22) | 31.3 (5) | .5224a | 25 (6) | 23 (13) | 23 (8) | .9537b |
| Loss of appetite | 15.8 (16) | 12.5 (2) | NSa | 25 (6) | 18 (10) | 5 (2) | .0908b |
| Nausea | 15.8 (16) | 6.3 (1) | .4613a | 16.7 (4) | 16 (9) | 11 (4) | .7381b |
| Rhinorrhoea | 10.9 (11) | 6.3 (1) | NSa | 4.2 (1) | 14 (8) | 8 (3) | .3430b |
| Pharyngeal pain | 5.9 (6) | 12.5 (2) | .3001a | 4.2 (1) | 9 (5) | 5 (2) | .6797b |
| Cough | 9.9 (10) | 6.3 (1) | NSa | 8.3 (2) | 11 (6) | 8 (3) | .8967b |
| Dyspnoea | 4 (4) | 6.3 (1) | .5273a | 8.3 (2) | 4 (2) | 3 (1) | .5332b |
| Muscle ache | 42.6 (43) | 68.8 (11) | .0620a | 33.3 (8) | 46 (26) | 54 (20) | .2839b |
| Joint pain | 27.7 (28) | 62.5 (10) | .0058b | 20.8 (5) | 30 (17) | 43 (16) | .1691b |
| Local reaction | |||||||
| Induration/nodule | 15.8 (16) | 12.5 (2) | NSa | 12.5 (3) | 16 (9) | 16 (6) | .9078b |
| Pain/sensitivity | 66.3 (67) | 75 (12) | .5771a | 66.7 (16) | 73 (41) | 59 (22) | .3806b |
| Rash | 5 (5) | 6.3 (1) | NSa | 4.2 (1) | 7 (4) | 3 (1) | .6043b |
| Oedema | 14.9 (15) | 6.3 (1) | .6942a | 8.3 (2) | 18 (10) | 11 (4) | .4345b |
| Itching | 5.9 (6) | 18.8 (3) | .1057a | 8.3 (2) | 11 (6) | 3 (1) | .3621b |
| Increased temperature | 20.8 (21) | 31.3 (5) | .3456a | 1.2 (3) | 27 (15) | 22 (8) | .3688b |
The data are presented as percentage (%) and absolute frequency. The significant values (P<.05) are displayed in bold.
This study was the first to investigate the association of the rs1800629 variants in TNF and rs2228145 in IL6R with the production of neutralising antibodies against SARS-CoV-2 and the frequency of ESAVI in the adult population of western Mexico who had been vaccinated with the AZD1222 vaccination regimen.
Insofar as the genotypic and allelic frequencies of the rs1800629 variants in the TNF gene and rs2228145 in the IL6R are concerned, the distribution coincides with what has been previously reported in the Mexican population.18,19 With respect to the percentages of neutralising antibodies, Chau et al found that the levels of neutralising antibodies against SARS-CoV-2 remained similar in both males and females.20 Furthermore, it has been reported that there is no difference between the levels of neutralising antibodies in vaccinated individuals compared to those who generated these antibodies following infection and that this response also fails to correlate with the sex of the subjects.21 In light of the aforementioned, our results are consistent with earlier research; nonetheless, it is important to note that the average number of days (36) post-vaccination in our participants was relatively short and this may account for the high percentages of neutralising antibodies detected against SARS-CoV-2 protein S (>90%). Neutralising antibody production against SARS-CoV-2, whether by natural infection or after vaccination with AZD12,21 tends to decline over time.
We did not find an association between the production of neutralising antibodies post-AZD1222 vaccination with the 2 genetic variants evaluated. As for the rs1800629 variant in TNF, there is nothing in the literature that has evaluated this relationship; however, Yao et al failed to establish an association between the rs1800629 variant and antibody production after immunisation with the Japanese encephalitis vaccine.15 Another study reported a correlation between the average level of antibodies and the presence of the rs1800629 variant after receiving the hepatitis B vaccine.14 Conversely, other authors have not reported an association between the rs1800629 variant and low antibody titres in individuals immunised with hepatitis B vaccine.22,23 As regards the rs2228145 variant in IL6R, there are no publications that have evaluated its association with the production of neutralising antibodies following immunisation with AZD1222. That said, a study in an Iranian population probed the link between the rs2228145 variant and the susceptibility to develop COVID-19, revealing that participants who carry the A allele in IL6R are more prone to develop COVID-19.24
In our study, the group of females vaccinated with AZD1222 had a higher frequency of all ESAVI, with the exception of restriction of movement. Evidence suggests that gender-based immunological, hormonal, genetic, and even gut microbiota differences might modulate the activity of the immune system, which could impact the outcome of the response to vaccination.25 In addition, it has been shown that following immunisation against the influenza, mumps, hepatitis A and B, smallpox, and dengue viruses, women develop more frequent and severe adverse reactions to vaccines such as fever, pain, and inflammation.25,26 Tran et al have determined that the most common ESAVI among those individuals who were immunised with AZD1222 were fever, muscle aches, fatigue, pain at the site of administration, and chills. Similar to our study, they found that the female group presented more frequent ESAVI compared to the male group.27 Another study reported no association between the frequency of ESAVI and sex after the first dose of AZD1222;28 however, it should be noted that the participants in our study received 2 doses of AZD1222.
When analysing the relationship between genetic variants in TNF and IL6R with the frequency of ESAVI after receiving the AZD1222 vaccine, the GA genotype of the rs1800629 variant in TNF was found to be associated with more frequent post-vaccination fever and joint pain, as well as a tendency to experience muscle ache more often in participants carrying this genotype. There are no previous reports with regard to this association; however, considering that TNF-α is a pro-inflammatory cytokine, the connection with these symptoms may be due to the physiological effects induced by TNF-α and possibly a higher expression and production of TNF-α in carriers of the GA genotype. This cytokine has been acknowledged as exerting physiological effects related to pain and temperature increase.29 Certain cytokines have the ability to induce prostaglandin synthesis in peripheral tissues, which could account for why this symptomatology tends to accompany the development of muscle ache and joint pain.30 Inasmuch as the effect of this genetic variant in the expression of the cytokine is concerned, the rs1800629 variant has been reported to elevate TNF gene expression;31 however, the evidence currently available is subject to deate.32 It is therefore important that the biological or functional effect of this variant be experimentally confirmed.
The limitations of this study include its sample size. Consequently, we recommend that the study be replicated in other populations and with a larger sample size; that other sociodemographic or anthropometric characteristics of the population be considered and their relationship with neutralisation percentages, as well as taking into account the days post-vaccination and quantifying serum levels of soluble TNF-α and IL-6R to determine whether they exhibit any changes after immunisation and if they correlate with the presence of ESAVI.
In conclusion, our study confirms a high percentage of neutralising antibodies generated against SARS-CoV-2 protein S in individuals who received the AZD1222 vaccine at an average of 36 (22–58) days post-vaccination. The ESAVI adynamia, chills, arthralgia, and pain at the site of administration displayed a higher incidence in the female group; but only fever and joint pain were associated with the rs1800629 variant in TNF in this Mexican population.
Ethical responsibilitiesThis research was approved by the Ethics Committee of the Centro Universitario del Sur of the University of Guadalajara, under registry code: CEI/046/2021 and by the Technical Research Committee, under registry code: SAC/CIP/019/2021. In addition, the principles laid down in the Declaration of Helsinki were complied with.
Informed ConsentAll the subjects in this studied participated voluntarily and signed the informed consent form.
FundingThis project was funded by the Centro Universitario del Sur de la Universidad de Guadalajara, in accordance with the Summons for the Enhancement of Academic Bodies, registration code: SAC/CIP/019/2021 awarded to Zyanya Reyes-Castillo.
Please cite this article as: Villa-Panduro AJ, Corona-Reynaga NM, Meza-Peña DA, Ramírez MAE, García ASE, Galindo-García J, Sanchez-Caballero B, Valdés-Miramontes EH, Muñoz-Valle JF, Reyes-Castillo Z. Genetic variants rs1800629 in TNF and rs2228145 in IL6R: Association with adverse event following immunisation (AEFI) and SARS-CoV-2 neutralising antibodies in western Mexico population that received AZD1222 vaccine. Vacunas. 2024. https://doi.org/10.1016/j.vacun.2023.12.004.