Non-Alcoholic Fatty Liver Disease (NAFLD) is a condition characterized by the accumulation of fat in the liver cells in individuals who consume little to no alcohol, often associated with metabolic syndrome, obesity, and insulin resistance. It ranges from simple steatosis to more severe forms like non-alcoholic steatohepatitis (NASH), which can lead to liver fibrosis, cirrhosis, and liver cancer. [1] Nonalcoholic fatty liver disease (NAFLD) is an increasing issue that affects approximately 25 % of the global population [2]. As the most common chronic liver disease in the world, NAFLD is highly related to obesity, metabolic syndrome, type 2 diabetes mellitus and cardiovascular disease [2]. However, approximately 10–20 % of patients with NAFLD have a normal body mass index (BMI), a noticeable subset we referred to as lean NAFLD [3]. Despite having a normal BMI, patients with lean NAFLD are considered metabolically unhealthy and have higher fasting glucose, insulin resistance, blood pressure, and dyslipidemia levels than lean controls [4]. Furthermore, lean NAFLD patients share a similar outcome with overweight-obese NAFLD patients in overall survival even if they do not eventually progress to overweight or obese [5]. Therefore, growing evidence indicates that lean NAFLD is a unique pathophysiological entity, but the aspects of clinical, histological and genetic heterogeneity are still unclear [6,7].
There are numerous gene studies to recognize lean NAFLD. Genome-wide association studies have indicated that single nucleotide polymorphisms (SNPs) in patatin-like phospholipase domain-containing 3 (PNPLA3) are the most extensively elucidated SNPs related to lean NAFLD [8,9]. PNPLA3 encodes a protein known as adiponutrin that is expressed in adipocytes and hepatocytes. Moreover, this protein has lipolytic and lipogenic properties that play a critical role in lipid metabolism [10]. The PNPLA3 rs738409 variant seemed to be more common in Japanese, Hong Kong and Sri Lankan patients with lean NAFLD than in those with obese NAFLD, but there was no difference in Western countries [11–13]. In addition, the presence of the PNPLA3 rs738409G allele has been associated with an earlier presentation of NAFLD in Taiwanese children [14]. Therefore, we hypothesized that PNPLA3 variants were independently associated with NAFLD as a risk factor for lean adults in the Taiwanese population. Sorting and assembly machinery component 50 homolog (SAMM50) and the well-known PNPLA3 are both located on chromosome 22q13. The SAMM50 gene encodes a protein that resides in the outer membrane of mitochondria and is involved in mitochondrial dynamics and biogenesis. Polymorphisms in SAMM50 have been associated with various metabolic diseases, such as non-alcoholic fatty liver disease (NAFLD) and obesity [15]. In Asian studies, SAMM50 polymorphisms contributed to the occurrence and severity of fatty liver in the Chinese Han, Korean and Japanese populations [16,17]. To date, no study has demonstrated the association of SAMM50 in lean subjects with and without NAFLD.
Accordingly, a case–control study was conducted in healthy and lean adults with and without NAFLD who underwent health exams and genotyping. We aimed to examine the different risks for lean NAFLD with and without PNPLA3 and SAMM50 variants in lean NAFLD patients compared with lean controls.
2Materials and Methods2.1Study populationThis was a case‒control study conducted in the HAVO Health Exam Clinic. The HAVO database included individuals aged more than 20 years who received a self-paid health check-up and SNP genotyping from Jan 2020 to the end of 2021. All subjects completed standardized questionnaires through individual interviews. The health survey questionnaire asked participants questions regarding socio-demographics, lifestyles and medical history. The health check-up included physical exams, blood analyses, and abdominal ultrasonography. Informed consent forms were signed. The study was approved by the Institutional Review Board of National Taiwan University Hospital (IRB NO. 202110005RIND).
For the lean NAFLD study population, the exclusion criteria were 1) body mass index ≥ 24 kg/m2; 2) excessive alcohol use, which was defined as drinking more than 30 g of alcohol daily for men and 20 g for women; 3) chronic liver diseases, which included chronic hepatitis, autoimmune, drug-induced, vascular, and inherited hemochromatosis, or Wilson disease. 4) inability to undergo an abdominal ultrasound examination; and 5) incomplete SNP genotyping. The content of the physical examination included weight and height, which were measured by a standard electronic scale and stadiometer, and blood pressure (BP), which was measured by an electronic sphygmomanometer. Waist circumference was measured horizontally through the middle point between the upper border of the iliac bones and the lower border of the ribs. The content of the blood test included fasting glucose, triglycerides and high-density lipoprotein cholesterol (HDL-C). Abdominal ultrasonography was performed by trained physicians. Fatty liver was binarily defined by the presence or absence of liver-kidney contrast. The Ministry of Health and Welfare in Taiwan has established a BMI cut-off of 24 kg/m² for overweight and 27 kg/m² for obesity recently [18,19]. We have consistently applied the BMI cut-off of 24 kg/m² in Taiwan to define lean and overweight-obese individuals. [20]. Participants were considered to have metabolic syndrome if they met ≥3 of the following criteria: waist circumference ≥90 cm in men or ≥80 cm in women; serum triglycerides ≥ 150 mg/dL; HDL-C < 40 mg/dL in men or <50 mg/dL in women; systolic BP ≥ 130 and/or diastolic BP ≥ 85 mm Hg; and fasting glucose ≥100 mg/dL. Participants with medications for diabetes, hypertension or hyperlipidemia were sorted into the group which met the criteria for fasting glucose ≥100 mg/dL, BP≥130/85 mmHg or serum TG ≥150 mg/dL respectively.
2.2Selection of single nucleotide polymorphismsWe searched candidate genes and NAFLD-related SNPs on the website of the most up-to-date SNPs reported in the publications of the National Human Genome Research Institute (NHGRI) and the European Bioinformatics Institute (EMBL-EBI) (Supplement A and B). The website of NHGRI-EBI Catalog of Published Genome-Wide Association Studies [21] collects a total of 67 documented SNPs in PNPLA3 and 12 SNPs in SAMM50 which were found to be related to NAFLD. From the HAVO database, a total of 1652 lean controls and 602 lean NAFLD patients were extracted by criteria. Global Screening Array-24 v1.0 BeadChip (Infinium, California, USA) was then used for genotyping. The basic microarray technical data of the Asian Screening Array were downloaded from the Illumina official website [22]. The main sources of ASA chips were from East Asian and Southeast Asian populations, such as China, Japan, South Korea, Mongolia, and Singapore. A total of more than 9000 subjects were enrolled, and whole-gene sequencing data were obtained. A total of approximately 642,824 SNPs were screened. The SNP genotyping panel for replication analysis was created using the Illumina VeraCode Genotyping assay. Data analyses of the custom panel results were conducted with Illumina GenomeStudio. The accuracy of the genotyping panel was verified using five duplicate samples with the Illumina HumanOmni-Quad BeadChip. Samples with a call rate below 0.99, SNPs with more than 2 % missing data, a Hardy-Weinberg P-value less than 0.01, or a minor allele frequency below 0.05 were excluded. For detailed methodology, please refer to previously published paper [23].
After matching 67 SNPs in PNPLA3 and 12 SNPs in SAMM50 to the ASA chip, we excluded 60 SNPs in the PNPLA3 gene and 8 SNPs in the SAMM50 gene. Then, we excluded one SNP for duplication and statistical non-significance and five SNPs in PNPLA3 and 2 SNPs in SAMM50 that had high collinearity (Pearson correlation coefficient > 0.95). As a result, rs738409 in PNPLA3 and rs3761472 in SAMM50 entered further statistical analyses. The flow chart of SNP selection for lean NAFLD is shown in Fig. 1.
Flow chart of SNP selection for lean NAFLD. Data was gathered from the HAVO Health Clinic cohort, which included 1652 lean controls and 602 lean NAFLD patients. Initially, 642,824 SNPs were identified using the Infinium GSA 24 v1.0 BeadChip. Significant SNPs were collected from the NHGRI-EBI Catalog of Published GWAS, resulting in the examination of 67 PNPLA3 and 12 SAMM50 SNPs. SNPs with duplications, non-significance, or high collinearity (Pearson correlation coefficient ≥ 0.95) were excluded. This process resulted in the final representative SNPs: rs738409 for PNPLA3 and rs3761472 for SAMM50. These representative SNPs were then used in the ROC analysis to evaluate their predictive value for lean NAFLD.
Data are presented as the mean±SD for continuous variables and number (percentage) for categorical variables. Differences between the groups were examined using the chi-squared test for categorical variables and Student's t-test or one-way analysis of variance (ANOVA) for continuous variables. Multivariate logistic regression analyses were performed to estimate the relationship between the odds of having lean NAFLD in relation to PNPLA3 rs738409 and SAMM50 rs3761472 after adjustment for age, sex, BMI and metabolic factors (waist circumference, fasting glucose, systolic BP, diastolic BP, triglycerides and HDL-C). We performed receiver operating characteristic (ROC) analysis to determine the diagnostic performance of PNPLA3 rs738409 and SAMM50 rs3761472 for lean NAFLD. All analyses were performed using SPSS statistical software (V.17, SPSS, Chicago, Illinois, USA) and R software (R-4.2.2). A p value of <0.05 was considered to statistical significance.
2.4Ethical statementInformed consent forms were signed, and the study was approved by the Institutional Review Board of National Taiwan University Hospital (IRB NO. 202110005RIND).
3Results3.1Baseline characteristicsA total of 1652 lean controls and 602 lean NAFLD patients were enrolled. The average age was 43.8 ± 11.5 years, and 1130 (50.1 %) patients were male (Table 1). In the lean NAFLD group, the subjects were older, and the percentage of metabolic syndrome was higher than that in the lean control group (case vs. control: 10.5 % vs. 1.5 %). Waist circumference, systolic BP, diastolic BP, fasting glucose and triglycerides were significantly higher and HDL-C was significantly lower in lean NAFLD patients than in lean controls.
Basic characteristics and biochemical profiles of the study population.
| Lean Control (n = 1652) | Lean NAFLD (n = 602) | P value | |
|---|---|---|---|
| Male (%) | 733 (44.4) | 397 (65.9) | <0.001 |
| Age (years) | 42.0 ± 11.4 | 48.6 ± 10.3 | <0.001 |
| Smoking (%) | 332 (20.4) | 202 (33.9) | <0.001 |
| BMI (kg/m2) | 21.0 ± 1.83 | 22.6 ± 1.1 | <0.001 |
| WC (cm) | 74.8 ± 6.3 | 81.4 ± 5.0 | <0.001 |
| SBP (mmHg) | 114.3 ± 12.1 | 119.5 ± 12.9 | <0.001 |
| DBP (mmHg) | 70.0 ± 9.1 | 74.3 ± 9.4 | <0.001 |
| Glu AC (mg/dL) | 87.5 ± 13.8 | 95.6 ± 24.1 | <0.001 |
| TCHO (mg/dL) | 190.6 ± 35.9 | 201.4 ± 34.7 | <0.001 |
| LDL (mg/dL) | 120.8 ± 34.0 | 134.7 ± 33.6 | <0.001 |
| TG (mg/dL) | 83.7 ± 40.9 | 136.5 ± 88.2 | <0.001 |
| HDL (mg/dL) | 59.0 ± 13.8 | 49.6 ± 11.7 | <0.001 |
| MetS (%) | 25 (1.5) | 63 (10.5) | <0.001 |
Abbreviations: BMI, body mass index; WC, waist circumference; SBP, systolic blood pressure; DBP: diastolic blood pressure; TCHO, total cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; HDL-C, high-density lipoprotein cholesterol.
The distribution of PNPLA3 rs738409 (CC/CG/GG) and SAMM50 rs3761472 (AA/AG/GG) in lean NAFLD patients differed significantly from that in lean controls. (Table 2A) In subjects with the PNPLA3 rs738409 CC/CG/GG genotypes, 193 (22.6 %), 216 (26.1 %) and 123 (36.1 %) had lean NAFLD compared with 660 (77.4 %), 611 (73.9 %) and 218 (63.9 %) lean controls, respectively [OR=1.11 (95% CI: 0.90–1.36) for the CG genotype, OR=1.77 (95% CI: 1.36–2.29) for the GG genotype (reference: CC genotype)]. In subjects with the SAMM50 rs3761472 AA/AG/GG genotypes, 197 (22.9 %), 287 (27.2 %) and 118 (35.0 %) had lean NAFLD compared with 664 (77.1 %), 769 (72.8 %) and 219 (65.0 %) lean controls, respectively [OR=1.26 (95% CI: 1.02–1.55) for the AG genotype, OR=1.82 (95% CI: 1.38–2.39) for the GG genotype (reference: AA genotype)] (data not shown).
Correlation between genotype at locus rs738409 (representative of PNPLA3 gene).
| CC (N = 853) | CG (N = 827) | GG (N = 341) | P value | |
|---|---|---|---|---|
| Male (%) | 421 (49.4) | 426 (51.5) | 160 (46.9) | 0.407 |
| Age (years) | 43.9 ± 11.6 | 43.3 ± 11.2 | 44.2 ± 11.5 | 0.362 |
| Smoking (%) | 201 (24.0) | 204 (24.8) | 75 (22.5) | 0.868 |
| BMI (kg/m2) | 21.4 ± 1.8 | 21.5 ± 1.8 | 21.4 ± 1.8 | 0.746 |
| WC (cm) | 76.5 ± 6.8 | 76.8 ± 6.7 | 76.4 ± 6.5 | 0.612 |
| SBP (mmHg) | 115.6 ± 12.6 | 115.8 ± 12.3 | 115.0 ± 12.4 | 0.771 |
| DBP (mmHg) | 71.2 ± 9.3 | 71.4 ± 9.5 | 70.8 ± 8.9 | 0.671 |
| Glu AC (mg/dL) | 89.3 ± 16.1 | 90.0 ± 17.7 | 89.8 ± 20.2 | 0.809 |
| TCHO (mg/dL) | 192.4 ± 34.1 | 194.1 ± 36.1 | 191.3 ± 39.0 | 0.544 |
| LDL (mg/dL) | 123.4 ± 32.4 | 126.1 ± 34.9 | 122.0 ± 36.5 | 0.116 |
| TG (mg/dL) | 99.4 ± 71.7 | 95.0 ± 55.5 | 96.5 ± 55.8 | 0.231 |
| HDL (mg/dL) | 56.5 ± 14.2 | 56.4 ± 13.5 | 56.5 ± 13.4 | 0.821 |
| MetS (%) | 37 (4.4) | 34 (4.1) | 11 (3.3) | 0.563 |
| Fatty liver (%) | 193 (22.6) | 216 (26.1) | 123 (36.1) | <0.001 |
We sorted the subjects into categories by genotype at locus rs738409 (representative of the PNPLA3 gene) or locus rs3761472 (representative of the SAMM50 gene). The rs738409 SNP posed a low, moderate and high risk of lean NAFLD in the CC, CG, and GG genotypes, respectively, and the rs3761472 SNP posed a low, moderate and high risk of lean NAFLD in the AA, AG, and GG genotypes, respectively. Among the different gene risks of fatty liver in the lean population, there were no significant differences in body mass index, age and sex. Additionally, none of the metabolic factors, including waist circumference, systolic BP, diastolic BP, fasting glucose, triglycerides and HDL-C, were significantly different among the different genotypes. The only difference among the CC, CG, and GG genotypes of PNPLA3 rs738409 or the AA, AG, and GG genotypes of SAMM50 rs3761472 was the presence or absence of lean NAFLD (Table 2B).
Correlation between genotype at locus rs3761472 (representative of SAMM50 gene).
| AA (861) | AG (N = 1056) | GG (N = 337) | P value | |
|---|---|---|---|---|
| Male (%) | 425 (49.4) | 545 (51.6) | 160 (47.5) | 0.354 |
| Age (years) | 43.8 ± 11.6 | 43.6 ± 11.3 | 44.2 ± 11.7 | 0.670 |
| Smoking (%) | 195 (23.0) | 266 (25.5) | 73 (22.2) | 0.330 |
| BMI (kg/m2) | 21.4 ± 1.8 | 21.5 ± 1.8 | 21.4 ± 1.7 | 0.896 |
| WC (cm) | 76.4 ± 6.7 | 76.7 ± 6.7 | 76.5 ± 6.5 | 0.554 |
| SBP (mmHg) | 115.6 ± 12.5 | 116.0 ± 12.6 | 115.0 ± 12.4 | 0.421 |
| DBP (mmHg) | 71.2 ± 9.3 | 71.3 ± 9.5 | 70.6 ± 8.9 | 0.509 |
| Glu AC (mg/dL) | 89.1 ± 16.1 | 90.2 ± 17.9 | 89.6 ± 20.0 | 0.443 |
| TCHO (mg/dL) | 192.4 ± 33.8 | 194.8 ± 36.4 | 191.9 ± 39.2 | 0.243 |
| LDL (mg/dL) | 123.3 ± 32.3 | 126.1 ± 35.3 | 122.8 ± 36.8 | 0.124 |
| TG (mg/dL) | 99.8 ± 72.2 | 97.3 ± 54.8 | 94.2 ± 54.8 | 0.353 |
| HDL (mg/dL) | 56.5 ± 14.2 | 56.4 ± 13.7 | 56.7 ± 13.5 | 0.935 |
| MetS (%) | 39 (4.6) | 41 (3.9) | 8 (2.4) | 0.225 |
| Fatty liver (%) | 197 (22.9) | 287 (27.2) | 118 (35.0) | <0.001 |
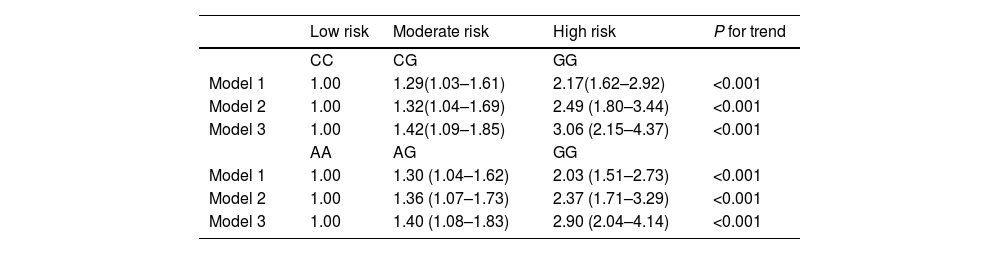
The odds ratios (ORs) of having fatty liver in lean subjects in relation to gene variants of PNPLA3 rs738409 and SAMM50 rs3761472 after adjustment by logistic regression models are shown in Table 3. For the PNPLA3 rs738409 SNP, the OR of having lean NAFLD for the GG group versus the CC group after adjustment for age, and sex was 2.17 (95% CI: 1.62–2.92; P for trend<0.001). After further adjustment for BMI and metabolic factors, i.e., waist circumference, fasting glucose, systolic BP, diastolic BP, triglycerides and HDL-C, the OR of having lean NAFLD for the GG group versus the CC group was 3.06 (95% CI: 2.15–4.37; P for trend <0.001). For the SAMM50 rs3761472 SNP, the OR of having lean NAFLD for the GG group versus the AA group after adjustment for age and sex was 2.03 (95% CI: 1.51–2.73; P for trend<0.001). After further adjustment for BMI and metabolic factors, the OR of having lean NAFLD for the GG group versus the AA group was 2.90 (95% CI: 2.04–4.14; P for trend <0.001).
The odds of having fatty liver in lean subjects in relation to PNPLA3 (rs738409) and SAMM50 (rs3761472) gene variants after adjustment by logistic regression models.
| Low risk | Moderate risk | High risk | P for trend | |
|---|---|---|---|---|
| CC | CG | GG | ||
| Model 1 | 1.00 | 1.29(1.03–1.61) | 2.17(1.62–2.92) | <0.001 |
| Model 2 | 1.00 | 1.32(1.04–1.69) | 2.49 (1.80–3.44) | <0.001 |
| Model 3 | 1.00 | 1.42(1.09–1.85) | 3.06 (2.15–4.37) | <0.001 |
| AA | AG | GG | ||
| Model 1 | 1.00 | 1.30 (1.04–1.62) | 2.03 (1.51–2.73) | <0.001 |
| Model 2 | 1.00 | 1.36 (1.07–1.73) | 2.37 (1.71–3.29) | <0.001 |
| Model 3 | 1.00 | 1.40 (1.08–1.83) | 2.90 (2.04–4.14) | <0.001 |
Model 1: adjusted for age and sex.
Model 2: adjusted for variables in Model 1 plus BMI.
Model 3: adjusted for variables in Model 2 plus metabolic factors: waist circumference, fasting glucose, systolic blood pressure, diastolic blood pressure, triglycerides and HDL-C.
Therefore, we used age, sex, waist circumference, fasting glucose, systolic BP, diastolic BP, triglycerides and HDL-C to calculate the area under the ROC for lean NAFLD in lean subjects. The AUCs were 0.859 (95%CI: 0.841, 0.877) and 0.860 (95%CI: 0.843, 0.877) for rs738409 and rs3761472, respectively (Fig. 2).
The area under the ROC curve for gene variants in the detection of lean NAFLD. Other than SNPs, sex, age, body mass index, waist circumference, systolic BP, diastolic BP, HDL and triglycerides were included in the analysis. The areas under the ROC curve for PNPLA3 rs738409 and SAMM50 rs3761472 in the detection of lean NAFLD were 0.859 (95%CI: 0.841, 0.877) and 0.860 (95%CI: 0.843, 0.877), respectively. A. PNPLA3 rs738409 and B. SAMM50 rs3761472.
In our study, we have identified gene variants of G alleles in PNPLA3 rs738409 and SAMM50 rs3761472 that are associated with a higher risk of lean NAFLD than in lean controls. In previous studies, there were inconsistent results for the association between PNPLA3 rs738409 and lean NAFLD, mostly due to ethnicity differences. Although SAMM50 has been linked to NAFLD, no study to date has explored this relationship. In this comprehensive cohort study, conducted on a large sample size, we observed a higher prevalence of the at-risk genotypes PNPLA3 rs738409 GG and SAMM50 rs3761472 GG in lean individuals with NAFLD compared to lean controls. These two variants, after adjustment for sex, age, BMI, fasting glucose, systolic BP, diastolic BP, triglycerides and HDL-C, had significantly higher odds ratios in lean NAFLD patients than in lean controls. The independent characteristics of PNPLA3 and SAMM50 for lean NAFLD, beyond BMI and metabolic syndrome, reconfirm the unique pathophysiology of lean NAFLD in Asian populations.
The variant PNPLA3 rs738409 was noted to be associated with NAFLD in a meta-analysis [10], among Asian populations [17] and in pediatric patients in Taiwan [14]. However, its association with lean subjects remains under debate in Western countries [2]. In Japanese studies, PNPLA3 rs738409 was strongly associated with the development and progression of nonobese NAFLD rather than obese NAFLD [24], which did not differ in metabolic morbidities and sex [13]. In Hong Kong, among those with NAFLD, lean subjects were more likely to carry the PNPLA3 rs738409 GG genotype than overweight and obese subjects [25]. Similarly, a biopsy-proven NAFLD population in China found that PNPLA3G and SAMM50 may synergistically interact to increase susceptibility to NASH [26]. Furthermore, patients with NAFLD who carry the PNPLA3G allele but are not obese have a higher risk of steatohepatitis or advanced fibrosis [11]. In line with a 7-year prospective community cohort in Sri Lanka, PNPLA3 variants showed higher associations with lean NAFLD [12]. Our study not only supported the higher association between the PNPLA3 rs738409 GG genotype and lean NAFLD in Taiwan but also demonstrated that the association was independent of BMI and metabolic dysfunction.
The role of SAMM50 in NAFLD is thought to be related to fatty acid oxidation and intracellular lipid accumulation [15]. Few studies have demonstrated the association between SAMM50 and NAFLD. In a Chinese Han population, the SAMM50 rs3761472G allele created susceptibility to NAFLD [27]. SAMM50 may interact with PNPLA3 to increase susceptibility to NAFLD in Chinese [28] and Mexican populations [29]. SAMM50 and PNPLA3 may affect the severity of NAFLD in Korean children [30] and adults [31] and the progression of NAFLD [16]. This was the first study to demonstrate that there was an independently higher risk of lean NAFLD among subjects with the SAMM50 rs3761472G allele variant after removing the effects of BMI and metabolic factors.
The 2024 EASL-EASD-EASO Clinical Practice Guidelines delineate the diagnostic criteria for Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD) [32]. Upon identifying hepatic steatosis through imaging or biopsy, the guidelines recommend evaluating the presence of cardiometabolic criteria. If the patient meets these cardiometabolic criteria and does not have a history of excessive alcohol consumption, the condition is classified as MASLD [33]. However, it is challenging to recognize whether a patient with normal body weight has fatty liver disease clinically. There is no reliable biomarker, and liver biopsy is not widely used for detecting lean NAFLD [34]. Our study demonstrated that PNPLA3 rs738409 or SAMM50 rs3761472 GG alleles have an approximately 3-fold higher risk of lean NAFLD after adjusting for BMI and metabolic factors. The area under the ROC curve (0.86) showed great performance for PNPLA3 rs738409 and SAMM50 rs3761472 in the detection of lean NAFLD.
There are some limitations of our study. First, it was a case‒control study where the exposure was PNPLA3 rs738409 or SAMM50 rs3761472 GG alleles and the outcome was lean NAFLD. Although we tried to collect and adjust for confounding factors such as age, sex, BMI, and every metabolic factor of MetS, potential confounders could not be ruled out. Second, we linked the PNPLA3 rs738409 and SAMM50 rs3761472 GG alleles to lean NAFLD and demonstrated an independent association of these variants with lean NAFLD. However, we could not distinguish the severity and progression of fatty liver with or without these genetic variants among lean subjects. Future studies should aim to explore the influences of polygenic genes in lean NAFLD. Additionally, our study design does not account for gene-environment interactions, which could also play a significant role in disease development.
5ConclusionsIn conclusion, the PNPLA3 rs738409 and SAMM50 rs3761472 gene polymorphisms are independently associated with a higher risk of fatty liver in lean individuals in Taiwan. Further validation of the association between lean NAFLD and genetic variants in Taiwan as well as Asian populations is warranted.
Author's contributionCWL, TJC and KCH designed the research; CWL, TJC, TYW, YHL, HJY and KCH conducted the research; CWL and TYW analyzed the data; CWL wrote the manuscript; CWL, TJC, TYW, YHL, HJY and KCH conducted the research; CWL and TYW revised the manuscript.
Availability of data and materialData will be made available on reasonable request
FundingThe study was partly funded by Min-Sheng General Hospital