In Colombia, between 2012 and 2013, 19 isolates with NDM were identified, of which 14 corresponded to Providencia rettgeri.
MethodsThe isolates were identified by Vitek-2, and antimicrobial susceptibility was evaluated by broth microdilution. The carbapenemase phenotypes were determined with Modified Hodge Test and synergy tests with EDTA/SMA and APB, the genotypes by PCR using specific primers for KPC, GES, IMP, VIM, OXA-48 and NDM, and genetic relationships were established with DiversiLab.
ResultsThe isolates were resistant to carbapenems, third-generation cephalosporins, piperacillin-tazobactam, amikacin, gentamicin and tigecycline, except aztreonam. All isolates were positive for EDTA/SMA and NDM-1, and negative for APB and other carbapenemases. Two genetic groups, group 1 (n=9 isolates), group 2 (n=4 isolates) and an isolate defined as not genetically related.
ConclusionThis work describes the circulating of NDM-1-producing P. rettgeri in Colombia.
En Colombia, entre 2012 y 2013 se identificaron 19 aislamientos con NDM, de los cuales 14 correspondían a Providencia rettgeri.
MétodosLos aislamientos se identificaron con Vitek-2, y la sensibilidad antimicrobiana se evaluó por microdilución en caldo. Las carbapenemasas se determinaron fenotípicamente con test de Hodge modificado y pruebas de sinergia con EDTA/SMA y APB, genotípicamente por PCR usando iniciadores específicos para KPC, GES, IMP, VIM, OXA-48 y NDM, y las relaciones genéticas se establecieron con Diversilab.
ResultadosLos aislamientos fueron resistentes a carbapenémicos, cefalosporinas de tercera generación, piperacilina-tazobactam, amicacina, gentamicina y tigeciclina, excepto a aztreonam. Todos los aislamientos fueron positivos para EDTA/SMA y NDM-1 y negativos para APB y otras carbapenemasas. Se definieron 2 grupos genéticos: grupo 1 (n=9 aislamientos), grupo 2 (n=4 aislamientos), y un aislamiento no relacionado genéticamente.
ConclusiónEste trabajo describe la circulación de grupos de P. rettgeri productores de NDM-1 en Colombia.
Providencia rettgeri is one of the most clinically significant species of the genus Providencia spp. This opportunistic pathogen causes multiple infections, such as: urinary tract infections, gastrointestinal infections and bacteraemia, among others.1 NDM was first described in January 2008, and this enzyme has spread rapidly worldwide, being detected mainly in enterobacteria and, to a lesser extent, in Acinetobacter spp. and Pseudomonas spp.2
In Colombia, NDM was first described in 2012, in isolates of Klebsiella pneumoniae that triggered an outbreak.3 From this report until December 2013, the National Institute of Health (INS) surveillance program for antimicrobial resistance in microorganisms causing healthcare-associated infections (HCAI), through the Microbiology Group (National Reference Laboratory), has confirmed 19 new cases of NDM (14 P. rettgeri, 3 Acinetobacter baumannii, one Pseudomonas aeruginosa and one K. pneumoniae, the latter a co-producer of NDM and KPC), observing that P. rettgeri NDM equals more than 70% of NDM-producing isolates in Colombia.
In consideration of the above, the objective of this report is to describe the microbiological and molecular characteristics of P. rettgeri NDM identified between 2012 and 2013 in Colombia.
Materials and methodsHistoryThe INS surveillance program for antimicrobial resistance in microorganisms that cause HCAI monitors various events of interest in public health, and includes the national laboratory surveillance of carbapenem resistance, which began in September 2012. From that date until December 2013, 749 isolates have been received (483 enterobacteria, 211 Pseudomonas spp. and 55 Acinetobacter spp.) for confirmation of carbapenemases, and these enzymes were detected in 529 (70.6%) isolates (343 enterobacteria, 136 Pseudomonas spp. and 50 Acinetobacter spp.). The most detected carbapenemase in enterobacteria was KPC (90.1%; n=309 isolates), in Pseudomonas spp. was VIM, followed by KPC (VIM: 70.6%; n=96 isolates, and KPC: 22.1%; n=30 isolates), and in Acinetobacter spp. was OXA-23 (86%; n=43 isolates); the cases of NDM in this period were 3.6% (n=19), and of these 73.7% (n=14) were P. rettgeri.
Bacterial isolatesThe INS Microbiology Group received 14 isolates of P. rettgeri, recovered in 5 hospitals of 3 cities located in 2 departments: hospital H1 (Bucaramanga-Santander), hospital H2 “outpatient centre” (Madrid-Cundinamarca) and hospitals H3-H5 (Bogotá D.C.-Cundinamarca).
Phenotypic characterisationIdentification at the genus and species level was confirmed using Vitek-2 (bioMérieux, Marcy l’Etoile, France) and antimicrobial susceptibility was assessed using broth microdilution (Sensititre panels; TREK Diagnostic Systems, Westlake, Ohio, USA). The results obtained were interpreted according to the parameters established by the CLSI,4 Except for tigecycline, for which the EUCAST criteria were used (breakpoints available at http://www.eucast.org/clinical_breakpoints/version 5.0). The phenotypic detection of carbapenemases was performed using: (a) modified Hodge test (MHT) (4), and (b) double-disc synergy tests using specific inhibitors for class B, carbapenemases (ethylenediaminetetraacetic acid/mercaptoacetic acid [EDTA/SMA]), and for class A, carbapenemases (Phenylboronic acid [PBA]).
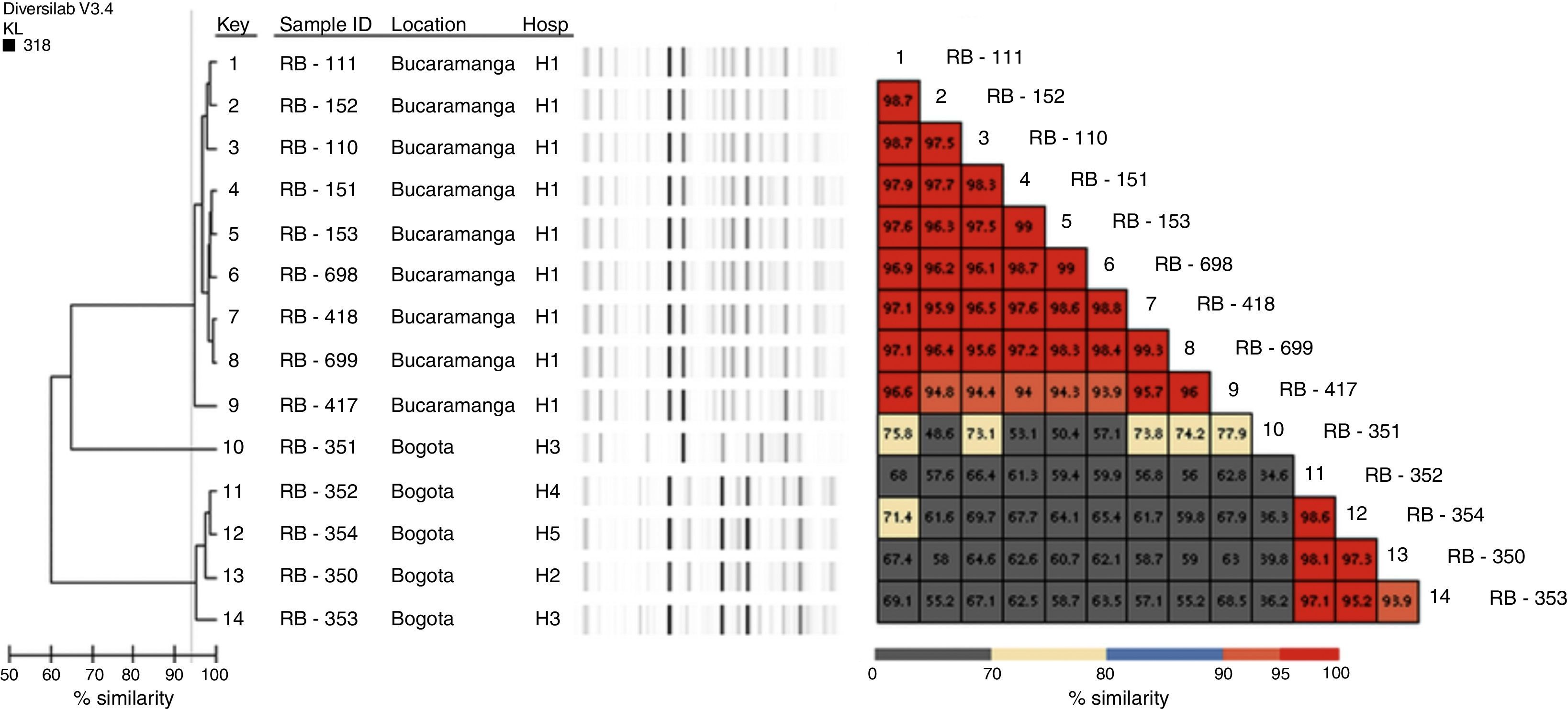
Genotypic characterisationThe detection of beta-lactamase encoding genes was performed by conventional PCR, for blaKPC, blaGES, blaVIM,5blaNDM,6blaIMP,7blaOXA-48,8blaCTX-M, blaTEM and blaSHV9; both chains of the carbapenemase amplification products were sequenced using an automated sequencer (Macrogen sequencing service, Korea, ABI PRISM® 3730XL from Applied Biosystem). Genotyping was performed using Diversilab (bioMérieux, Marcy l’Etoile, France) using the bacterial kit, and the results were analysed with the Kullback Leibler correlation (based on the presence and absence of bands rather than intensity); isolates with similarity >95% were considered indistinguishable; with similarity of 93–95%, closely related, and with similarity <93%, different.
ResultsBacterial isolatesP. rettgeri was recovered in patients ranging in age from 16 to 88years, being primarily recovered in men (85.7%; n=12) and in urine samples (71.4%; n=10) associated with both infection and colonisation (Table 1). Nine isolates came from the H1 hospital, most of them classified as colonisation (n=6), isolated in different departments of the institution, between 2012 and 2013 (Table 1). The remaining 5 isolates were isolated in hospitals H2 to H5 in 2013, with one isolation per institution, except H3, which had 2 isolates (Table 1).
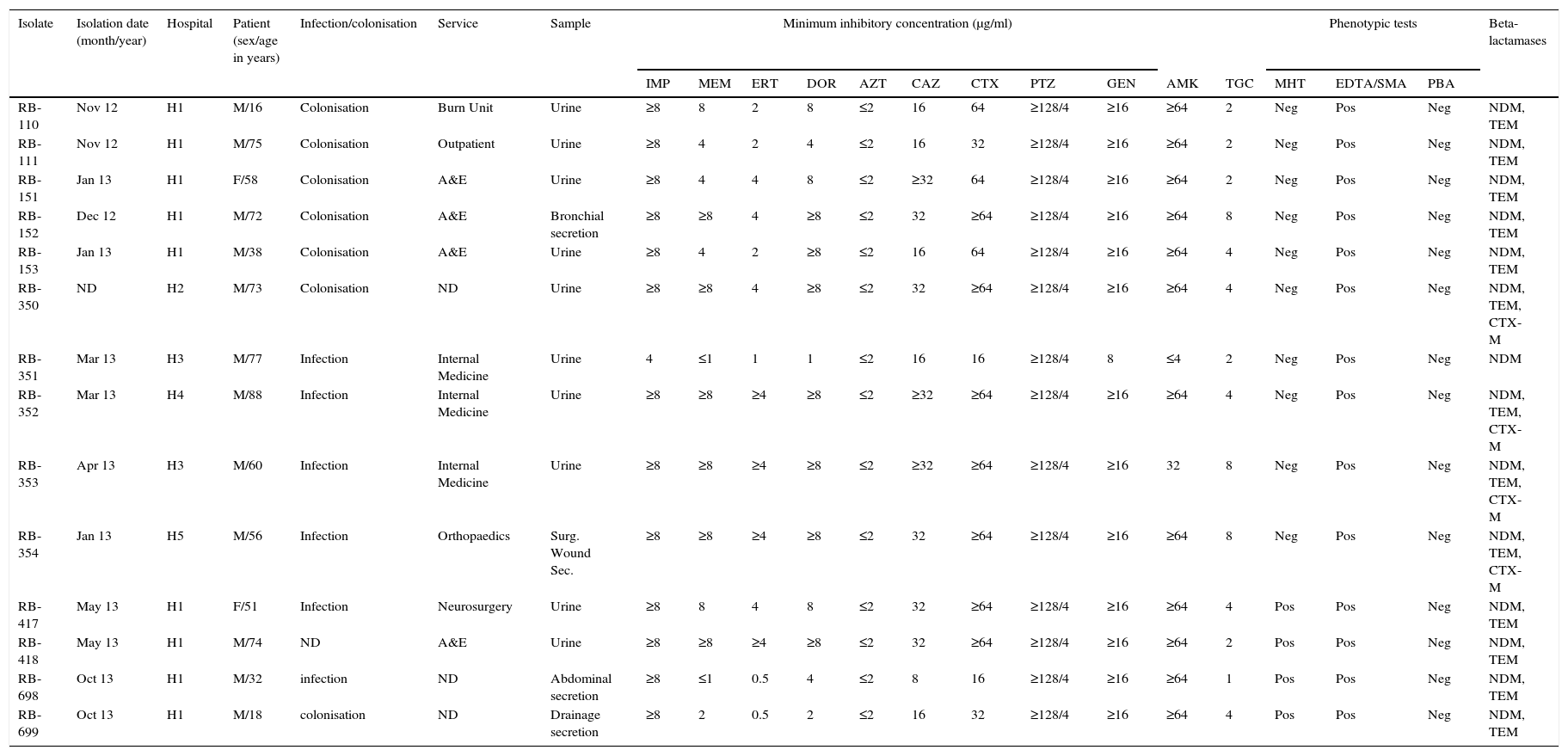
Clinical, phenotypic and genotypic characteristics of the isolates.
| Isolate | Isolation date (month/year) | Hospital | Patient (sex/age in years) | Infection/colonisation | Service | Sample | Minimum inhibitory concentration (μg/ml) | Phenotypic tests | Beta-lactamases | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | ERT | DOR | AZT | CAZ | CTX | PTZ | GEN | AMK | TGC | MHT | EDTA/SMA | PBA | ||||||||
| RB-110 | Nov 12 | H1 | M/16 | Colonisation | Burn Unit | Urine | ≥8 | 8 | 2 | 8 | ≤2 | 16 | 64 | ≥128/4 | ≥16 | ≥64 | 2 | Neg | Pos | Neg | NDM, TEM |
| RB-111 | Nov 12 | H1 | M/75 | Colonisation | Outpatient | Urine | ≥8 | 4 | 2 | 4 | ≤2 | 16 | 32 | ≥128/4 | ≥16 | ≥64 | 2 | Neg | Pos | Neg | NDM, TEM |
| RB-151 | Jan 13 | H1 | F/58 | Colonisation | A&E | Urine | ≥8 | 4 | 4 | 8 | ≤2 | ≥32 | 64 | ≥128/4 | ≥16 | ≥64 | 2 | Neg | Pos | Neg | NDM, TEM |
| RB-152 | Dec 12 | H1 | M/72 | Colonisation | A&E | Bronchial secretion | ≥8 | ≥8 | 4 | ≥8 | ≤2 | 32 | ≥64 | ≥128/4 | ≥16 | ≥64 | 8 | Neg | Pos | Neg | NDM, TEM |
| RB-153 | Jan 13 | H1 | M/38 | Colonisation | A&E | Urine | ≥8 | 4 | 2 | ≥8 | ≤2 | 16 | 64 | ≥128/4 | ≥16 | ≥64 | 4 | Neg | Pos | Neg | NDM, TEM |
| RB-350 | ND | H2 | M/73 | Colonisation | ND | Urine | ≥8 | ≥8 | 4 | ≥8 | ≤2 | 32 | ≥64 | ≥128/4 | ≥16 | ≥64 | 4 | Neg | Pos | Neg | NDM, TEM, CTX-M |
| RB-351 | Mar 13 | H3 | M/77 | Infection | Internal Medicine | Urine | 4 | ≤1 | 1 | 1 | ≤2 | 16 | 16 | ≥128/4 | 8 | ≤4 | 2 | Neg | Pos | Neg | NDM |
| RB-352 | Mar 13 | H4 | M/88 | Infection | Internal Medicine | Urine | ≥8 | ≥8 | ≥4 | ≥8 | ≤2 | ≥32 | ≥64 | ≥128/4 | ≥16 | ≥64 | 4 | Neg | Pos | Neg | NDM, TEM, CTX-M |
| RB-353 | Apr 13 | H3 | M/60 | Infection | Internal Medicine | Urine | ≥8 | ≥8 | ≥4 | ≥8 | ≤2 | ≥32 | ≥64 | ≥128/4 | ≥16 | 32 | 8 | Neg | Pos | Neg | NDM, TEM, CTX-M |
| RB-354 | Jan 13 | H5 | M/56 | Infection | Orthopaedics | Surg. Wound Sec. | ≥8 | ≥8 | ≥4 | ≥8 | ≤2 | 32 | ≥64 | ≥128/4 | ≥16 | ≥64 | 8 | Neg | Pos | Neg | NDM, TEM, CTX-M |
| RB-417 | May 13 | H1 | F/51 | Infection | Neurosurgery | Urine | ≥8 | 8 | 4 | 8 | ≤2 | 32 | ≥64 | ≥128/4 | ≥16 | ≥64 | 4 | Pos | Pos | Neg | NDM, TEM |
| RB-418 | May 13 | H1 | M/74 | ND | A&E | Urine | ≥8 | ≥8 | ≥4 | ≥8 | ≤2 | 32 | ≥64 | ≥128/4 | ≥16 | ≥64 | 2 | Pos | Pos | Neg | NDM, TEM |
| RB-698 | Oct 13 | H1 | M/32 | infection | ND | Abdominal secretion | ≥8 | ≤1 | 0.5 | 4 | ≤2 | 8 | 16 | ≥128/4 | ≥16 | ≥64 | 1 | Pos | Pos | Neg | NDM, TEM |
| RB-699 | Oct 13 | H1 | M/18 | colonisation | ND | Drainage secretion | ≥8 | 2 | 0.5 | 2 | ≤2 | 16 | 32 | ≥128/4 | ≥16 | ≥64 | 4 | Pos | Pos | Neg | NDM, TEM |
AMK: amikacin; PBA: synergism test with phenylboronic acid inhibitor; CAZ: ceftazidime; CTX: cefotaxime; DOR: doripenem; EDTA/SMA: synergism test with inhibitor (ethylenediaminetetraacetic acid/mercaptoacetic acid EDTA/SMA; ERT: ertapenem F: female GEN: gentamicin IMP: imipenem M: male Internal med.: internal medicine MEM: meropenem; Neg: negative; Pos: Positive: PTZ: piperacillin-tazobactam; ND: no data; Surg. Wound Sec: Surgical wound secretion; Sec: secretion; TGC: tigecycline; MHT: modified Hodge test; Burn U: Burn unit.
The isolates were resistant to all antimicrobials evaluated; carbapenems, third generation cephalosporins, piperacillin-tazobactam, amikacin, gentamicin and tigecycline, except aztreonam (Table 1). All isolates were positive for EDTA/SMA, negative for PBA, and only 4 isolates were positive for MHT (Table 1).
Genotypic characterisationAll isolates were positive for NDM-1, 13 for TEM and 4 for CTX-M (Table 1). No isolate amplified KPC, GES, IMP, VIM, OXA-48 or SHV. Classification with Diversilab helped define 2 clonal groups of 9 (group 1) and 4 (group 2) isolates, respectively (Fig. 1), and one isolate that is not genetically related to the rest. Group 1 is composed of 9 isolates (8 indistinguishable and one closely related), all from the H1 hospital, while group 2 consists of 3 indistinguishable isolates and one closely related from hospitals H2 to H5 (Fig. 1). All isolates of group 2 were positive for CTX-M.
DiscussionIn Colombia, various types of carbapenemases are circulating, and it is considered an endemic region for KPC carbapenemases, detected mainly in enterobacteria and Pseudomonas spp.,10,11 VIM mainly identified in Pseudomonas spp.,11 and OXA-23 in Acinetobacter baumannii.12 Although in Colombia NDM was detected just under 5 years ago,3 currently its identification has increased in the country according to INS surveillance data (these data are not shown), which is a matter of great concern due to the rapidity of dissemination of this carbapenemase worldwide and the possibility that in our country this mechanism of resistance may become of high prevalence, as has happened with other carbapenemases.
In Latin America, NDM was detected for the first time in isolates of Klebsiella pneumoniae in Guatemala13 and Colombia3; at present this enzyme has been isolated in different gram-negative microorganisms in the region, including P. rettgeri, which has been identified in Argentina, Brazil, Mexico and Uruguay,13 and this is the first report in Colombia. Early detection of carbapenemases in the microbiology laboratory through the use of screening tests is important for preventing the dissemination of these resistance determinants in the hospital setting. In this study it was observed that the identification of class B carbapenemases (NDM) using the test with EDTA/SMA was 100% sensitive and specific, whereas MHT only managed to detect less than 30% of these isolates, which is in agreement with that reported in the literature.2,4
The majority of microorganisms producing carbapenemases have a multiresistance profile, limiting the treatment options, which is why colistin has become one of the antimicrobials of choice to treat these infections.2 However, in the case of P. rettgeri with NDM, colistin cannot be considered as treatment due to the natural resistance of this microorganism.14 In this study, the majority of isolates presented a multiresistance phenotype and were only sensitive to aztreonam; this characteristic is being studied as a possible strategy to treat these patients by using a combination of aztreonam and avibactam (betalactamase inhibitor NXL104).2,15
This study describes the dissemination of 2 clonal groups; one at the intrahospital level where P. rettgeri NDM occurred in an institution in Bucaramanga (H1) and another where P. rettgeri NDM and CTX-M were identified in 4 institutions (3 of Bogotá and one of Madrid [city located 28km from Bogotá]). In Colombia the occurrence of an outbreak by isolates with NDM had been previously described.3 However, this is the first time that the spread of a clonal group has been described in several institutions, and although no relationship was established between patients, it is important to establish permanent and articulated epidemiological and laboratory surveillance in order to take control measures to prevent dissemination of NDM isolates in institutions in these geographical areas.
In conclusion, in Colombia the circulation of 2 genetic groups of P. rettgeri NDM-1 is observed. Therefore, it is important to strengthen surveillance at a laboratory level to detect these enzymes, which will allow the necessary measures to be taken to control the spread of these determinants of resistance at the hospital level.
Conflicts of interestThe authors declare that they have no conflicts of interest.
To the Departmental Public Health Laboratories (LDSP) of Cundinamarca (Amparo Leonor Génecco Rodríguez), the LDSP of Santander (Leonor Chacón de Mendieta) and the District Health Secretary of Bogotá (María del Socorro Chala). The HCAI Team (healthcare-associated infections). Subdirectorate of Surveillance and Control. Surveillance monitoring and risk management. National Institute of Health. To the institutions, Hospital Universitario de Santander E.S.E., Hospital El Tunal E.S.E., Hospital la Victoria E.S.E., Hospital Simón Bolívar E.S.E. and Centro Ambulatorio Gustavo Escallón Cayzedo – Fundación Santa Fe de Bogotá.
Please cite this article as: Saavedra-Rojas S-Y, Duarte-Valderrama C, González-de-Arias M-N, Ovalle-Guerro MV. Emergencia de Providencia rettgeri NDM-1 en dos departamentos de Colombia, 2012-2013. Enferm Infecc Microbiol Clin. 2017;35:354–358.