Despite conducting studies to investigate food contamination in hospitals in different parts of Iran in recent years, there have been no reliable studies to identify Salmonella Enteritidis, Salmonella Typhimurium, Bacillus cereus, Bacillus subtilis, and Clostridium perfringens in hospital food in Mashhad. Therefore, this study was conducted with the aim of investigating some major foodborne pathogens in hospital food. In this study, 360 food samples were randomly selected from 12 different menus from 13 hospitals affiliated with Mashhad University of Medical Sciences, Mashhad, Iran. Microbial culture methods for the recovery/isolation or enumeration of Salmonella spp., Bacillus spp. and C. perfringens as well as toxinotyping of C. perfringens using the PCR method were performed. B. cereus and C. perfringens were detected in 4 out of 360 food samples, 2 (0.55%) of which were B. cereus and, the remaining 2 (0.55%) were C. perfringens; B. subtilis was not detected in any of the food samples. Furthermore, Salmonella was found in 21 (5.82%) food samples, 12 (3.33%) of which were S. Typhimurium, 4 (1.11%) were S. Enteritidis, and 5 (1.38%) belonged to other Salmonella species. The most contaminated foods were salad, kebab, and rice samples, which accounted for 36%, 16%, and 12% of the contaminated foods, respectively. In our study, two strains of S. Typhimurium and S. Enteritidis, were the primary causative agents of food contamination among the investigated pathogens. More stringent control measures should be implemented in hospital catering, particularly for unprocessed foods such as salads.
A pesar de que en los últimos años se han realizado algunos estudios para investigar la contaminación de alimentos en hospitales de diferentes partes de Irán, no se cuenta con datos confiables referidos a Salmonella Enteritidis, Salmonella Typhimurium, Bacillus cereus, Bacillus subtilis y Clostridium perfringens en alimentos hospitalarios en Mashhad, Irán. Este estudio tuvo por objetivo investigar algunos de los principales patógenos transmitidos por alimentos en alimentos hospitalarios. Se seleccionaron al azar 360 muestras de alimentos de 12 menús diferentes de 13 hospitales de la Universidad de Ciencias Médicas de Mashhad. Se realizaron métodos de cultivo microbiano para la recuperación, el aislamiento y el recuento de Salmonella spp., Bacillus spp. y C. perfringens. Se tipificaron las toxinas de este último microorganismo mediante el método de PCR. B. cereus se detectó en 2 muestras (0,55%) y C. perfringens en otras 2 muestras. B. subtilis no se detectó en ninguna de las muestras. Además, se encontró Salmonella en 21 (5,82%) muestras, donde 12 (3,33%) fueron S. Typhimurium, 4 (1.11%) fueron S. Enteritidis y 5 (1.38%) fueron otras especies de Salmonella. Los alimentos más contaminados fueron la ensalada, el kebab y las muestras de arroz, que constituyeron el 36%, el 16% y el 12% de los alimentos contaminados, respectivamente. Dos cepas de S. Typhimurium y S. Enteritidis causaron la mayor contaminación de alimentos entre los patógenos investigados. Deben seguirse medidas de control más estrictas en la alimentación hospitalaria, especialmente en alimentos no procesados, como las ensaladas.
Food safety and quality control as priorities throughout the supply chain are attracting more attention every day and is indeed an important aspect of public health. Although there is an advanced monitoring system in most countries worldwide, the occurrence of waterborne and foodborne diseases continues47. In the United States, approximately 9 million cases of foodborne illness occur each year, resulting in approximately 120000 hospitalizations and 3000 deaths22. In Iran, the outbreak rate of foodborne diseases was 1.38 cases/100000 population in 2011. The Hazard Analysis of Critical Control Points (HACCP) system, along with the standards set by the International Organization for Standardization (ISO), is one of the most efficient methods for ensuring food safety in production and distribution centers in Iran. The HACCP system is used to identify potential risk points and wrong practices in production and processing processes. Moreover, a foodborne disease surveillance system, along with a national guideline for foodborne diseases, was developed by the Center for Communicable Disease Control6,16.
In general, foodborne diseases are caused by the consumption of food or water contaminated with pathogens or their toxins23,24. Pathogens that cause foodborne diseases are often referred to as foodborne pathogens, and include bacteria, viruses, fungi, and parasites13,37,42. There are 31 identified foodborne pathogens, and it is estimated that viruses are the primary causes of illnesses, whereas bacteria are the primary causes of hospitalizations and deaths14,31. The most prevalent pathogens that cause foodborne illnesses include Listeria monocytogenes, C. perfringens, Escherichia coli, Staphylococcus aureus, Salmonella spp., Shigella spp., Campylobacter jejuni, Clostridium botulinum, Cronobacter sakazakii, B. cereus, Vibrio spp., and Yersinia enterocolitica, viruses (Hepatitis A and Noroviruses), parasites (Cyclospora cayetanensis, Toxoplasma gondii, and Trichinella spiralis)21,44.
Salmonella species are one of the most common foodborne pathogens and a public health concern worldwide, causing significant economic consequences1,9,28. Gastroenteritis is the most common Salmonella infection in humans and, is caused by Salmonella serotypes, especially S. Typhimurium and S. Enteritidis. Acute gastroenteritis, typhoid enteric fever, paratyphoid fever, and systemic infections are caused by S. Typhimurium and S. Enteritidis serotypes12,35,36. These two serotypes are recognized as the most important intestinal pathogens in animals and humans that are transmitted through contaminated water and food, and are considered to be a health problem worldwide33,36,48. It is estimated that 11 million cases of typhoid fever, 1.3 million cases of gastroenteritis, and 3 million deaths worldwide are annually attributable to Salmonella27. The primary transmission route for Salmonella infections is fecal-oral, which involves consuming food or drink that has been tainted with the patient's feces or urine. Contaminated soils, sediments, and water can also play an important role in the transmission of Salmonella38. Eggs, poultry, and dairy products are the most common sources of Salmonella. In recent years, fresh products such as fruits and vegetables have also been suggested as a method of transmission of the infection40,52.
B. cereus is one of the most important causes of food poisoning in humans. This pathogen is found in various foods, such as rice and cereals, as well as meat and dairy products, and has been identified as an opportunistic pathogen. Some isolates of B. cereus can grow under refrigeration, and its spores can withstand very high temperatures18. B. cereus is responsible for two different types of food poisoning: diarrhoea and vomiting. Diarrheal disease has an incubation period of 8–16h and causes abdominal pain and distinct diarrhoea. This disease is caused by enterotoxins produced during the growth of B. cereus in the small intestine. Another type of disease is characterized by nausea and vomiting and is caused by a vomiting toxin produced by vegetative cells in food, a condition that has an incubation period of 0.5–5h17.
C. perfringens causes disease in humans and livestock. In addition, this bacterium is abundantly present in soil, water, food, faeces, and the intestines of humans and animals36,39,41. C. perfringens is capable of producing at least 17 toxins, all of which are proteins. The four main toxins, alpha (α), beta (β), epsilon (&#¿;), and uta (ι) are used to classify C. perfringens into types A, B, C, D, and E10,50,51. C. perfringens type A is found in the soil of contaminated areas and is not found in the intestines of healthy animals. Type C causes stroke and necrotic enteritis in piglets, lambs, calves, and humans. Type D causes enterotoxicity in lambs and, sheep. Type E is also the cause of hemorrhagic enteritis in calves, and although rare, it has been reported in Australia19. Among these types, the epsilon toxin is one of the most important toxins of this bacterium7. Enterotoxins caused by C. perfringens are a common cause of food poisoning in humans29.
Because different types of Salmonella spp., Bacillus spp., and C. perfringens cause different diseases in humans, it is therefore essential to investigate their presence in hospital catering services.
Materials and methodsStudy designAll hospitals in Mashhad city were classified into four categories: academic, private, military, and charitable. Among these four categories, 13 university hospitals in Mashhad city were selected for evaluation and sampling.
From 12 shared menus including grilled dishes (chicken kubideh, chicken barbecue, kubideh), meat dishes (lentil pilaf with minced meat, vegetable pilaf with fish, fried chicken pilaf, Istanbuli with meat), stewed dishes (Gheimeh, Gormeh saezi), rice (bean pilaf, rice), and salad; 360 food samples were randomly taken from 13 academic hospitals in Mashhad in three intervals and in triplicate. The collected food samples were stored in sterilized glass jars and kept in ice chests and transported to the laboratory under aseptic conditions (0°C, 15min).
The food analysis is provided by the hospital canteen to researchers and officials at the educational hospitals in Mashhad. This allows them to conduct additional studies in the future and add their findings to the results of this study to further identify contaminated hospital foods and take effective actions to reduce the contamination of food pathogens. These results are aimed both at the sick and the general public.
Microbiological analysisSalmonellaPre-enrichment, bacterial enrichment, and selective media are necessary for the detection of Salmonella strains.
- •
First step: For pre-enrichment of Salmonella, 225g of a peptone water buffer environment for 24h at 37°C.
The samples were prepared, diluted, and subjected to microbiological analysis in compliance with the guidelines provided by the Institute of Standards and Industrial Research of Iran with respect to the total count enumeration of Salmonella (No. 1810, identical to ISO 6579-1: 2017).
- •
Second step: For selective enrichment, 1ml of the pre-enriched samples in BPW was inoculated into a tube containing 9ml of cysteine selenite culture medium and tetrathionate broth (SCB and TTB, Liofilchem, Roseto Degli Abruzzi, Italy). The tube was then kept at 43°C for 24h.
- •
Third step: The cultures were streaked onto Salmonella-Shigella agar (Merck, Darmstadt, Germany) and xylose lysine deoxycholate agar (Merck) and incubated at 37°C for 24–48h. Black-centered colorless colonies were identified as Salmonella and tested by culturing in triple sugar iron agar (TSIA, Merck). After incubation at 37°C for 24h, hydrogen sulphide gas, the black and yellow color at the bottom, and the red color in the slant of the TSI tubes were observed.
- •
Fourth step: Colony identification was confirmed by the PCR assay based on the detection of Salmonella spp. S139–S140 gene, and the results were recorded based on the presence/absence of Salmonella spp25,32.
First step: After preparing the initial suspension, 0.1ml was cultured on the surface of MYP (Mannitol egg yolk polymyxin) medium and incubated at 35°C for 24–48h.
Second step: B. cereus colonies were identified by counting large pink colonies with a whitish precipitate zone. Using a PCR assay based on the identification of the B. cereus GroEL gene, colony identification was verified2.
C. perfringensFirst step: 25ml of prepared serial dilution was enriched in perfringens enrichment medium (PEM, Liofilchem, Roseto Degli Abruzzi, Italy). For this, 25ml of prepared serial dilution was mixed with 225ml of PEM and incubated anaerobically at 45°C for 24h.
Second step: the samples were prepared in tryptose sulphite cycloserine agar (TSCA, Liofilchem, Roseto Degli Abruzzi, Italy), without egg yolk (TSC), and, incubated at 37°C for 24h under anaerobic conditions in a gas pack system (Gas Pak, BBL, Merck, Darmstadt, Germany)45.
Third step: For the purposes of anaerobic morphological and biochemical identification, the black colonies on the TSCA were chosen. Colony identification was verified using a PCR assay based on the detection of the C. perfringens 16S rDNA gene.
Fourth step: Vegetative colonies were typed by PCR because of the presence of genes responsible for toxin production (alpha, beta, epsilon, and uta) and virulence genes (tpeL) and (netB)26.
DNA extractionTo extract DNA, the boiling method was used. Two bacterial colonies were used for this purpose and were boiled at 100°C for 10min in a water bath inside a tube that held 50μl of distilled water. Following a 5-min centrifugation at 13000rpm, 5μl of the supernatant was transferred into a different sterile tube with a capacity of 2ml. Using a nanodrop spectrophotometer (model: 2000 UV-Vi, Thermo Fisher Scientific, Deutschland, Germany), the concentration of the extracted DNA was determined and used for PCR2.
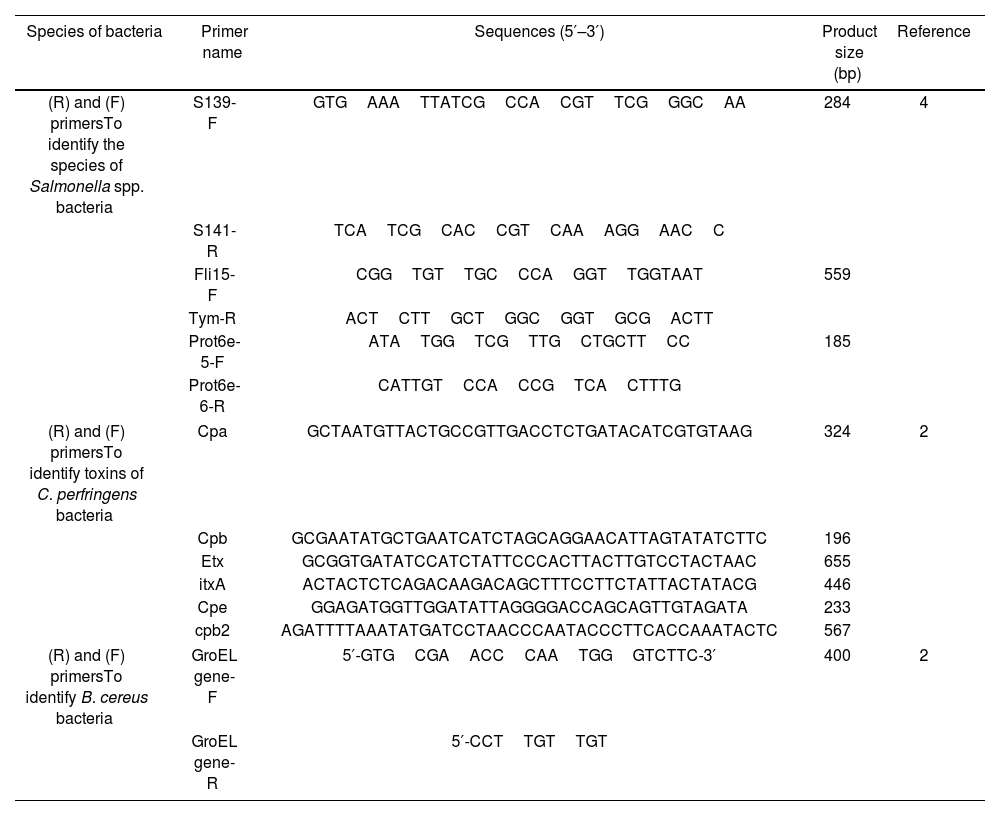
Performing multiplex PCRThe PCR assay used typical colonies on selective media as templates. These colonies were verified by biochemical tests. Sterile distilled water served as the negative control, whereas Salmonella spp. (PTCC-1609), C. perfringens (ATTC-1312), and B. cereus (ATCC-10876) were used as positive controls. The study primers are displayed in Table 1.
Primers used in the PCR.
| Species of bacteria | Primer name | Sequences (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|---|
| (R) and (F) primersTo identify the species of Salmonella spp. bacteria | S139-F | GTGAAATTATCGCCACGTTCGGGCAA | 284 | 4 |
| S141-R | TCATCGCACCGTCAAAGGAACC | |||
| Fli15-F | CGGTGTTGCCCAGGTTGGTAAT | 559 | ||
| Tym-R | ACTCTTGCTGGCGGTGCGACTT | |||
| Prot6e-5-F | ATATGGTCGTTGCTGCTTCC | 185 | ||
| Prot6e-6-R | CATTGTCCACCGTCACTTTG | |||
| (R) and (F) primersTo identify toxins of C. perfringens bacteria | Cpa | GCTAATGTTACTGCCGTTGACCTCTGATACATCGTGTAAG | 324 | 2 |
| Cpb | GCGAATATGCTGAATCATCTAGCAGGAACATTAGTATATCTTC | 196 | ||
| Etx | GCGGTGATATCCATCTATTCCCACTTACTTGTCCTACTAAC | 655 | ||
| itxA | ACTACTCTCAGACAAGACAGCTTTCCTTCTATTACTATACG | 446 | ||
| Cpe | GGAGATGGTTGGATATTAGGGGACCAGCAGTTGTAGATA | 233 | ||
| cpb2 | AGATTTTAAATATGATCCTAACCCAATACCCTTCACCAAATACTC | 567 | ||
| (R) and (F) primersTo identify B. cereus bacteria | GroEL gene-F | 5′-GTGCGAACCCAATGGGTCTTC-3′ | 400 | 2 |
| GroEL gene-R | 5′-CCTTGTTGT | |||
The multiplex PCR test was performed in a final volume of 25μl containing 12.5μl PCR Premix (Taq DNA Polymerase 2x Master Mix Red includes 0.4mmol dNTP, tris–hydrochloric acid, ammonium sulphate, 3mmol MgCl2 (1.5mM), 0.2% Tween 20, 0.2 units per μl Taq DNA polymerase amplicon, red color, and stabilizer) (AMPLIQON company), 1μl each primer (R and F) (0.2μM, AMPLIQON company), 5.5μl sterile deionized distilled water, and 5μl of extracted DNA. A thermocycler (Model BIORAD T100, US) was used to program the amplification.
All pairs of primers (Salmonella sp., serotypes Enteritidis, S.) Typhimurium were added to a single PCR reaction in these reactions, along with the previously prepared reagent mix. The reaction contained primers that amplified Enteritidis S139-F and S141-R genes (284bp) as well as Typhimurium (Fli15-F, Tym-R, 559bp) (Prot6e-5-F, Prot6e-6-R, 185bp) (Table 1). The amplification reaction and cycling conditions were as follows: initial incubation at 95°C for 5min, 35 cycles of denaturation at 94°C for 1min, annealing at 56°C for 30s, extension at 72°C for 1min, and a final extension period of 10min at 72°C4.
To determine the toxin typing of Clostridium isolates using the multiplex PCR technique, the nucleotide sequence of the primers used is shown in Table 1 to ascertain the presence of the cpa, cpb, itxA, etx, cpe, and cpb2 genes. Two strains of C. perfringens 1312-ATCC from the Department of Food Hygiene, Faculty of Veterinary Medicine, Ferdowsi University of Mashhad were used as positive controls to determine the presence of the cpa, cpb, itxA, etx, cpe, and cpb2 genes. The initial cycle was set for 5min at 95°C, and there were 40 subsequent cycles at 94°C for 1min, 56°C for 1min, 72°C for 1min, and a final extension at 72°C for 5min5.
For B. cereus, the initial denaturation of the partially synthesized DNA was performed at 95°C for 5min. Thereafter, 30 cycles of amplification were performed, with denaturation at 94°C for 30s, annealing at 63°C for 30s, an extension at 72°C for 30s, and a final extension at 72°C for 5min.
The PCR amplicons were examined using agarose gel electrophoresis, in which 5μl of the PCR products were loaded onto 1.5% agarose gels containing 0.45g agarose powder mixed with 30ml of TAE buffer X1 and 1μl of Greenweaver. Then, electrophoresed for 35–45min at 100–120V5. The PCR products were then observed under UV radiation, and the images of the bands corresponding to the sequences amplified by the PCR reaction were noted and preserved (Model Gel documentation, Uvite).
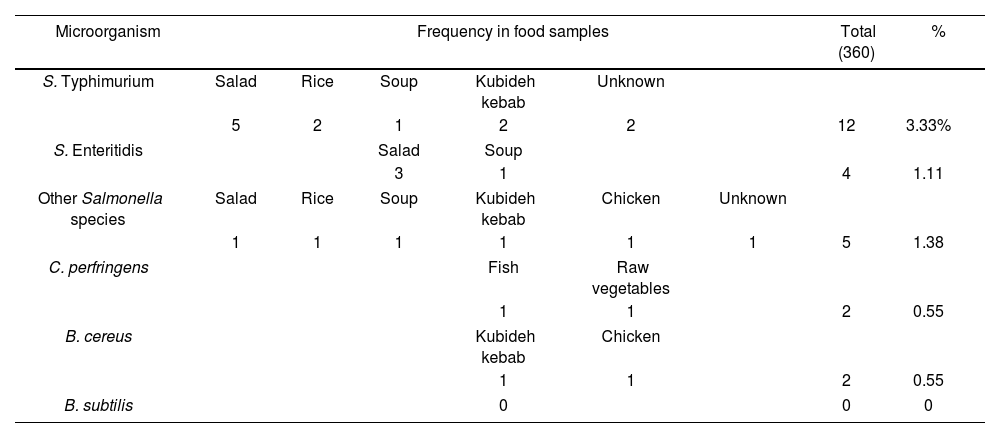
ResultsThree hundred and sixty hospital food samples were examined. In these 360 food samples, two cases of C. perfringens were observed; none of these two instances included toxin-producing strains. Two food samples included B. cereus, but none contained B. subtilis. Finally, Salmonella was found in 21 food samples; 12 were S. Typhimurium, 4 were S. Enteritidis, and 5 belonged to other Salmonella species. The items mentioned are shown in Table 2. Figure 1 shows the amplified genes of B. cereus, S. Typhimurium, and C. perfringens.
Pathogens found in hospital food menus.
| Microorganism | Frequency in food samples | Total (360) | % | |||||
|---|---|---|---|---|---|---|---|---|
| S. Typhimurium | Salad | Rice | Soup | Kubideh kebab | Unknown | |||
| 5 | 2 | 1 | 2 | 2 | 12 | 3.33% | ||
| S. Enteritidis | Salad | Soup | ||||||
| 3 | 1 | 4 | 1.11 | |||||
| Other Salmonella species | Salad | Rice | Soup | Kubideh kebab | Chicken | Unknown | ||
| 1 | 1 | 1 | 1 | 1 | 1 | 5 | 1.38 | |
| C. perfringens | Fish | Raw vegetables | ||||||
| 1 | 1 | 2 | 0.55 | |||||
| B. cereus | Kubideh kebab | Chicken | ||||||
| 1 | 1 | 2 | 0.55 | |||||
| B. subtilis | 0 | 0 | 0 | |||||
PCR results on agarose gel electrophoresis. (a) Amplified GroEL genes with band length of 400bp related to B. cereus. Well No. 2 is the positive control, Well No. 3 is the negative control, Well No. 5 and 4 are samples. (b) Amplified genes of fliC with a band length of 559bp and Prot6e with a band length of 185bp related to Salmonella bacteria. Well number 2 from the left is the positive control, well number 3 is the negative control, wells number 5–16 are samples of S. Typhimurium and 17–21 are other Salmonella species. (c) Amplified InvA genes with a band length of 284bp related to S. Enteritidis bacteria. Well number 2 from the left side is the positive control, well number 3 is the negative control, wells number 4–7 are samples. (d) Amplified 16S rDNA genes with a band length of 279bp related to C. perfringens. Well number 2 is sample and positive control, number 3.
Table 2 examines the type of food in which each of the mentioned pathogens was found. Based on this, most cases of S. Typhimurium (5 cases) and S. Enterica (3 cases) were detected in salad. Moreover, S. Typhimurium was found in rice (2 cases), kubideh (2 cases), and soup (1 case). In addition to the salad sample, S. Enteritidis was found in soup (1 case). Other Salmonella species were found in salad (1 case), rice (1 case), chicken (1 case), and kubideh kebab (1 case). C. perfringens was found in fish (1 case), and raw vegetable (1 case). In addition, B. cereus was found in salad (1 case) and kebab (1 case). The food items that were most contaminated with the investigated pathogens included salad, kubideh kebab, and rice.
DiscussionFoodborne diseases have been introduced as an essential health problem in different countries. Considering that foodborne diseases can be much more severe and dangerous in high-risk people such as children and infants, pregnant women and their fetuses, and people who have a weak immune system, identifying risk factors is an important issue, and as a result, people need more health protocols. Some foodborne pathogen symptoms that have been reported in immunocompromised patients include persistent diarrhea, Norovirus (NoV) infection, which is frequently waterborne or foodborne and increases the risk of serious consequences, including chronic gastroenteritis. Hepatitis E infection in immunocompromised organ transplant patients, invasive infections, chronic hepatitis, or hepatitis E8,34. In the present study, we identified the species of S. Enteritidis, S. Typhimurium, B. cereus, and B. subtilis and determined the toxin types of C. perfringens in packaged hospital foods in Mashhad hospitals.
The results obtained from our study showed that 7.5% of the examined hospital foods were contaminated with different types of Salmonella, of which almost half were S. typhi. C. perfringens was present in 1% of the samples, although none of the strains found were toxin-containing. Finally, B. cereus was also found in two food samples, but no case of B. subtilis was observed. The foods that were most contaminated with the investigated pathogens included salad, kubideh kebab, and rice.
In a study conducted in 2017, Zahedi et al. investigated the contamination of raw red and white meat with S. Typhimurium and S. Enteritidis in Shahrekord city, Iran. The obtained results showed that the highest amount of Salmonella contamination was related to chicken meat, beef, goat meat, and sheep meat, and the highest contamination of all Salmonella isolates examined was S. Typhimurium and S. Enteritidis, respectively. Similarly, in our study, both S. Typhimurium and S. Enteritidis were among the factors contributing to food contamination. In contrast, our study examined cooked foods, whereas Zahedi et al. examined raw meats.
In a study conducted by Deilmi et al.11, enterotoxicogenic B. cereus was identified in prepared meat foods in Zanjan city, Iran. The presence of B. cereus was tested in 80 different meat samples (sausages, hamburgers, and ready-made kebabs) purchased from stores in Zanjan City. Finally, out of the 80 examined samples, 32 samples were infected with B. cereus. In our study, only two samples of B. cereus contamination were found; one was in grilled meat and the other was in chicken. In any case, because our study was conducted on hospital foods, perhaps the lower prevalence of contamination found in our study can be attributed to greater compliance with hygiene principles in hospital kitchens compared with the meat preparation centers available on the market.
The majority of food service staff in hospitals possess good food safety knowledge, practice, and attitude. Passing some food hygiene courses by food service staff plays a crucial role in the enhancement of their function in the hospitals. Some measures were implemented for the hospital staff, such as regular handwashing before handling food, not using the same cutting board for raw and cooked foods, storing raw food separately from cooked food, ensuring cooked meat is thoroughly checked using thermometers, thawing frozen food in the refrigerator or other cool place, washing fruit and vegetables with safe water, and cleaning surfaces and equipment used for food preparation before reusing food. On the other hand, food handlers in some hospitals can play a role in increasing food contamination due to general ignorance of pathogenic agents, improper storage temperatures for cold and hot ready-to-eat foods, a lack of food hygiene training, and inadequate performance in the area of food safety3,24,30.
In another study conducted in 2016 by Deilami et al.10, in Zanjan, the authors isolated B. cereus from 220 food samples, including 80 prepared meat products, 20 dairy samples, including pasteurized and local milk, cheese, cream, and ice cream, 20 rice samples, and 100 salad samples served in Tabriz and Zanjan restaurants. The obtained results showed that 100 of the studied food samples were contaminated with B. cereus. The level of contamination with B. cereus in the hospital foods examined in our study was much lower than that in this study, which may be related to greater compliance with hygiene in hospital canteens, as previously mentioned. It is also possible that the epidemiological differences in the prevalence of pathogens in Mashhad and Zanjan caused this difference in the obtained results.
In general, foodborne diseases emerge more strongly in hospitalized patients. Therefore, food hygiene in hospitals is important and significant. Improper temperature storage, improper cooking, insufficient cooling of food, long-term food storage process, infectious diseases of food service center employees, and use of contaminated tools and equipment are the factors that increase the risk of foodborne illness for people in the hospital. To reduce the prevalence of contamination in hospitals, there are several recommendations and management strategies, including using appropriate tools for monitoring food services. HACCP is one of these powerful tools to ensure and implement proper safety conditions in catering. This system describes the features and conditions for selecting raw materials and other necessary facilities for transportation, storage, processing, packaging, distribution, and consumption. It also provides guidelines for the provision of food for vulnerable groups by accredited suppliers who comply with legal requirements. Preventive measures include promoting personal hygiene and proper handling practices to avoid the transfer of pathogens from personnel to consumers, ensuring sufficient cooking and proper storage temperature control, preventing secondary contamination, installing air filters against airborne contamination, and enhancing the knowledge and awareness of food handlers15,20,43,46,49.
ConclusionMost food contamination in our study was caused by various species of Salmonella, such as S. Typhimurium and S. Enteritidis, out of all the pathogens found in hospital food in Mashhad, Iran. B. cereus and C. perfringens contamination levels were relatively low. In this study, the food samples did not contain any B. subtilis. In terms of food contamination, rice, kubideh kebab, and salad contained the highest levels of the pathogens analyzed.
FundingMashhad University of Medical Science Ethics Committee granted its clearance for this study (970405).
Conflict of interestNo conflict of interest declared.
This study was supported by the Research project number 970405 in Mashhad University of Medical science, Mashhad, Iran.