The objective is to develop a model that predicts vital status six months after fracture as accurately as possible. For this purpose we will use five different data sources obtained through the National Hip Fracture Registry, the Health Management Unit and the Economic Management Department.
Material and methodsThe study population is a cohort of patients over 74 years of age who suffered a hip fracture between May 2020 and December 2022. A warehouse is created from five different data sources with the necessary variables. An analysis of missing values and outliers as well as unbalanced classes of the target variable (“vital status”) is performed. Fourteen different algorithmic models are trained with the training. The model with the best performance is selected and a fine tuning is performed. Finally, the performance of the selected model is analysed with test data.
ResultsA data warehouse is created with 502 patients and 144 variables. The best performing model is Linear Regression. Sixteen of the 24 cases of deceased patients are classified as live, and 14 live patients are classified as deceased. A sensitivity of 31%, an accuracy of 34% and an area under the curve of 0.65 is achieved.
ConclusionsWe have not been able to generate a model for the prediction of six-month survival in the current cohort. However, we believe that the method used for the generation of algorithms based on machine learning can serve as a reference for future works.
Desarrollar un modelo que prediga con la mayor exactitud posible el estado vital a los 6 meses tras fractura de cadera, utilizando para ello cinco fuentes de datos obtenidas a través del Registro Nacional de Fracturas de Cadera, la Unidad de Gestión Sanitaria y la Dirección Económica.
Material y metodologíaLa población de estudio es una cohorte de pacientes que sufrieron fractura de cadera entre mayo de 2020 y diciembre de 2022. A partir de cinco fuentes diferentes de datos se crea un almacén con las variables necesarias. Se realiza un análisis de valores perdidos y atípicos, así como de desbalanceo de las clases de la variable objetivo («estado vital»). Se entrenan 14 diferentes modelos algorítmicos con los datos de entrenamiento. Se selecciona el modelo que mejor rendimiento obtenga y se realiza una puesta a punto fina. Finalmente se analiza el rendimiento del modelo con datos de test.
ResultadosSe crea un almacén de datos con 502 pacientes y 144 variables. El modelo con mejor rendimiento es la regresión lineal. Dieciséis de los 24 casos de pacientes fallecidos son clasificados como vivos, y 14 pacientes vivos son clasificados como fallecidos. Se consigue una sensibilidad del 31%, una precisión del 34% y un área bajo la curva de 0,65.
ConclusionesNo se ha conseguido generar un modelo de predicción de muerte a los 6 meses con nuestra cohorte. Sin embargo, creemos que el método utilizado para generar algoritmos basados en aprendizaje automático puede servir de referencia para futuros trabajos.
Machine learning tools have not only revolutionised healthcare technologies,1 but their applications can also be used on medical data to increase the power of traditional statistical analyses.2 In addition, current electronic medical data management systems rely on different sources of data (medical records, analyses, electrocardiograms, radiological images, etc.). These sources individually provide excellent information for the day-to-day management of our patients. But the possibility of combining or merging various data sources opens the door to new possibilities for analysis and research.3
Supervised learning is a type of machine learning in which the data offered to the machine is labelled (for example, in this paper the label is the vital status of the patient, alive/dead), so it has to generate an algorithm capable of classifying the data correctly according to these labels. A supervised model, therefore, is a mathematical formula, or algorithm, that the machine uses to attempt to classify the data correctly. Examples of these supervised models include the simplest, such as logistic regression, or the more complex, such as the Gradient Boosting Classifier.
In our case, we performed an analysis of different supervised machine learning models to determine their ability to predict the vital status (alive/dead) of a cohort of patients over 74 years old at 6 months after a hip fracture. The aim was to obtain a model that can accurately predict vital status 6 months after fracture using only data that can be retrieved during hospital admission. For this purpose, we used five different data sources obtained through the National Hip Fracture Registry (RNFC), the Health Management Unit (UGS), and our hospital's financial department.
Material and methodsFig. 1 shows a schematic of the database preparation process and the strategy for supervised model generation.
Preparation of the databaseThe data sources used, the method used to add them to a data warehouse, as well as the procedure used to generate the supervised mortality prediction models are described below.
Data sources (Appendix)In order to test the ability to merge databases from different sources, we decided to select variables which, a priori, could be related to the event studied (vital status at 6 months) and which are generated throughout the hospital stay of the hip fracture patient.
RNFC. RNFC is a multicentre registry of the epidemiological, clinical, functional, and care characteristics of patients with hip fracture and follow-up at one month after hospital discharge in several hospitals in Spain.4 The present study uses a cohort corresponding to the cases registered in our hospital from May 2020 to December 2022. We selected patients discharged alive after hospital admission for hip fracture. Of all the variables collected in the registry, we used the 40 obtained during hospital admission.
Presbide. Presbide is the electronic prescription system of the Basque Health Service (Osakidetza). It is considered a priori that both the number and type of prescription may be related to patient survival (for example, lower survival may be assumed in patients prescribed a loop diuretic, such as furosemide, by the internist, as it is usually indicated in patients with congestive heart failure). The UGS provided information on the prescriptions prescribed at hospital discharge after admission for hip fracture. We grouped the most frequent brands of medicines into 17 groups, and also selected other medicines which, although outside these 17 groups, had been prescribed ten or more times. We also created the variable ‘Total number of prescriptions’, which is the sum of these variables.
Vital status at 6 months. The UGS provided the vital status (alive/dead) of the patients as of 31 June 2023, as well as the date of death, if such an event had occurred. The calculation was made to determine the vital status of the patient 6 months after admission for hip fracture.
Implants. Given the possibility that the type of implant may be related to higher or lower survival of the patients (for example, patients prescribed a total hip prosthesis may have higher survival than those implanted with a bipolar prosthesis), information was requested from the financial department about the implants used during the surgical intervention of patients with hip fracture. The implant subtype, the implant family, and the specific implant model are specified. All subtypes and families were selected, as well as the specific model of the implants that had been implanted on 20 or more occasions. Finally, we made a total count of all implants registered for each patient.
Total price of the implants. The financial department also provided information on the total price excluding VAT of the materials implanted in each patient during hip fracture surgery, in case there could be any causal relationship (for example, the higher the price of the implant, the greater the complexity of the surgery, and the lower the survival rate).
Extraction, transformation, and load (ETL)Taking into account the heterogeneity of both the data and the sources used, an information extraction procedure from the sources was required. This extraction involves transforming, cleaning, enriching, and integrating the data to finally load them into a data warehouse. A NoSQL database was created for these processes using the open source database system MongoDB. The final product was a data warehouse in which, in addition to the variables described above, other variables are stored that may be useful for subsequent analysis of this cohort.
Pre-processing strategies: handling of missing values, outliers, and unbalancingThe missing values in the database were analysed and the imputation model chosen according to the nature and characteristics of the missing values, ranging from case elimination to imputation with other values (a constant value, a measure of central tendency such as the mean, or more complex techniques such as imputation with K-Nearest Neighbours).
Potential outliers were visualised using principal component analysis (PCA). If outliers were identified, a database with outlier elimination was created using the interquartile range rule (IQR).5 Provided that the number of outliers removed does not exceed 10% of the total number of records, this transformed database was used.
The target variable of the supervised models analysed was ‘vital status’ at 6 months after admission for hip fracture, which is a dichotomous variable (‘alive/dead’). If either class totals equal or are more than 90% of the cases, the database is considered to be unbalanced, and therefore it is balanced using the Synthetic Minority Over-sampling Technique (SMOTE),6 which generates synthetic examples of the minority class to balance the class distribution.
Supervised model generationOnce cleaned and prepared, the database was split at a 70:30 ratio into two parts: one for training the models (training data, 70%) and one for testing the performance of the models (test data, 30%).
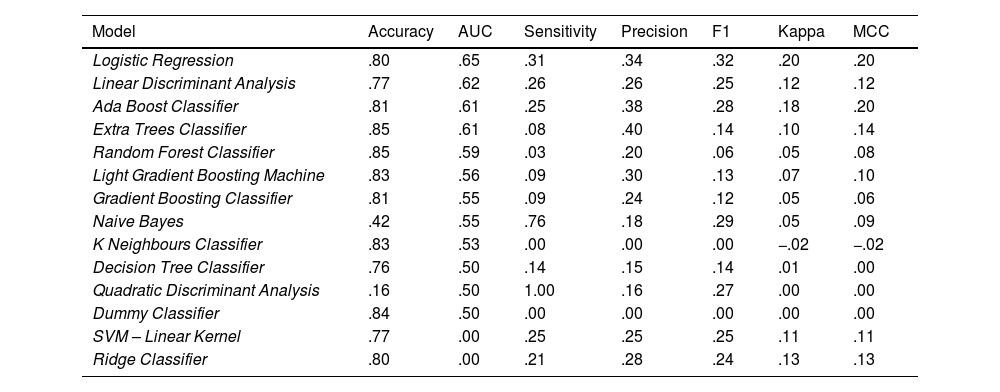
Thirteen different algorithmic models (Table 1), plus a reference ‘dummy classifier’, were trained on the training data following a 10K-folds cross-validation strategy (the training data were divided into 10 groups, so that the models were trained 10 times, taking 9 out of these 10 groups in each iteration). These 14 trained models were tested with the test data and 7 performance parameters were calculated (Table 1).
List of models and performance parameters used.
| Supervised models analysed | Performance parameters used |
|---|---|
| Ada Boost Classifier | Accuracy |
| Decision Tree Classifier | AUC |
| Dummy Classifier | Sensitivity |
| Extra Trees Classifier | Accuracy |
| Gradient Boosting Classifier | F1 |
| K Neighbours Classifier | Kappa index |
| Light Gradient Boosting Machine | Matthews correlation coefficient (MCC) |
| Linear Discriminant Analysis | |
| Naive Bayes | |
| Quadratic Discriminant Analysis | |
| Random Forest Classifier | |
| Ridge Classifier | |
| SVM – Linear Kernel |
The model with the best performance in the area under the curve (AUC) was selected and fine tuned by modifying its parameters, in 10 cross-validation tests using a segmentation of 10K-folds (in total 100 adjustments). If better performance was obtained, the best of these fine-tuned models was selected.
Finally, the ROC (Receiver Operating Characteristic) curve of the selected model was generated, in addition to the confusion matrix, and the weight of the different variables in the model construction analysed.
Description of the tools usedThe NoSQL MongoDB database was implemented on the NoSQLBooster administration tool and worked in Python code through the Pymongo module.
The process of ETL was carried out with the open-source Pandas library.
The database pre-processing, the generation of supervised models and the performance analysis were carried out with the Pycaret module.
The Python code work with the Pymongo, Pandas, and Pycaret modules was implemented in the Jupyter Notebook application.
All data analysis work was done in local settings to avoid traffic and leakage of non-anonymised data over the Internet.
ResultsThe process of ETL was carried out, obtaining a database of 502 patients, whose characteristics are shown in Table 2. Only 9 of the 144 variables used have missing values, in percentages of less than 1.2% (Table 3). These are mainly cases of patients who were treated conservatively, so that some of the data, such as those of the surgical intervention, are missing. Taking into account that these are polynomial qualitative variables and that in the RNFC code 11 was used in cases where the information cannot be retrieved, a constant value, code 12, was imputed for these missing values.
Characteristics of the study population.
| Mean age in years | 87.13 |
| % Female sex | 74.6 |
| % ASA risk (II/III/IV) | 20.3/55.7/24 |
| % Right laterality | 49.1 |
| Type of fracture (%) | Non-displaced subcapital: 5.3 |
| Displaced subcapital: 40.2 | |
| Pertrochanteric: 46.5 | |
| Subtrochanteric: 8 | |
| Type of surgery (%) | Conservative: .2 |
| Cannulated: 1 | |
| Sliding screw: 14.3 | |
| Intermedullary nail: 40.1 | |
| Cemented hemiarthroplasty: 40.1 | |
| Total prosthesis: 4.5 | |
| % death at 6 monthsa | 19.2 |
| Surgical delay in hours | 48.87 |
| Hospital stay in days | 12.05 |
Variables with missing values and their percentages (%).
| Variables with missing values | Percentage of missing values |
|---|---|
| Non-weightbearing after surgery | 1.2 |
| Osteoprotective treatment at discharge | .6 |
| Antiresorptive at discharge | .6 |
| Bone former at discharge | .6 |
| Calcium at discharge | .6 |
| Vitamin D at discharge | .6 |
| Other treatments at discharge | .6 |
| Anaesthetic block | .4 |
Potential outliers are observed in the PCA analysis, especially in cases of patients who survived beyond the sixth month after admission for hip fracture (Fig. 2). The elimination of outliers leads to the loss of only 18 cases (from 502 to 484), which is 3.58% of the total, and therefore we decided to perform the analysis with elimination of outliers.
The resulting database after the elimination of outliers shows 76 cases of death, compared to 408 cases of living patients. Although there is an imbalance between the two classes (15.7% versus 84.3%), it does not reach 90%, and therefore no technique was used to correct it.
Generation of supervised modelsThe most accurate model is Random Forest, with 85% of cases well classified. For the AUC parameter, linear regression obtains a value of .65. Linear regression also obtains the best value for F1 (.32), the Matthews correlation coefficient (MCC; .2), and the kappa index (.2). The quadratic discriminant analysis has 100% sensitivity, although the values for the other performance parameters are poor (e.g. 50% accuracy and an AUC of .5, which means it is making random predictions). The Extra Tree Classifier is the most accurate, with 40% of really deceased patients well classified among the death predictions.
Although the Dummy Classifier has good accuracy (84%), the AUC and the other parameters indicate that its predictions are random.
As for the results of other models, the poor performance of the Gradient Boosting Classifier is striking, with an AUC of .55 and an F1 of .12 (Table 4).
Performance parameters of the 14 supervised models analysed.
| Model | Accuracy | AUC | Sensitivity | Precision | F1 | Kappa | MCC |
|---|---|---|---|---|---|---|---|
| Logistic Regression | .80 | .65 | .31 | .34 | .32 | .20 | .20 |
| Linear Discriminant Analysis | .77 | .62 | .26 | .26 | .25 | .12 | .12 |
| Ada Boost Classifier | .81 | .61 | .25 | .38 | .28 | .18 | .20 |
| Extra Trees Classifier | .85 | .61 | .08 | .40 | .14 | .10 | .14 |
| Random Forest Classifier | .85 | .59 | .03 | .20 | .06 | .05 | .08 |
| Light Gradient Boosting Machine | .83 | .56 | .09 | .30 | .13 | .07 | .10 |
| Gradient Boosting Classifier | .81 | .55 | .09 | .24 | .12 | .05 | .06 |
| Naive Bayes | .42 | .55 | .76 | .18 | .29 | .05 | .09 |
| K Neighbours Classifier | .83 | .53 | .00 | .00 | .00 | −.02 | −.02 |
| Decision Tree Classifier | .76 | .50 | .14 | .15 | .14 | .01 | .00 |
| Quadratic Discriminant Analysis | .16 | .50 | 1.00 | .16 | .27 | .00 | .00 |
| Dummy Classifier | .84 | .50 | .00 | .00 | .00 | .00 | .00 |
| SVM – Linear Kernel | .77 | .00 | .25 | .25 | .25 | .11 | .11 |
| Ridge Classifier | .80 | .00 | .21 | .28 | .24 | .13 | .13 |
The model with the best AUC performance is linear regression. The original model is fine tuned, but none of the 100 adjustments analysed obtains better results, and therefore we decided to continue working with the original linear regression model (see parameters in Table 5).
Configuration of the selected linear regression model.
| (‘trained_model’, | |
| LogisticRegression (C=1.0, class_weight=None, | |
| dual=False, | |
| fit_intercept=True, | |
| intercept_scaling=1, | |
| l1_ratio=None, max_iter=1000, | |
| multi_class=‘auto’, n_jobs=None, | |
| penalty=‘l2’, random_state=231, | |
| solver=‘lbfgs’, tol=.0001, | |
| verbose=0, | |
| warm_start=False))], | |
| verbose=False) |
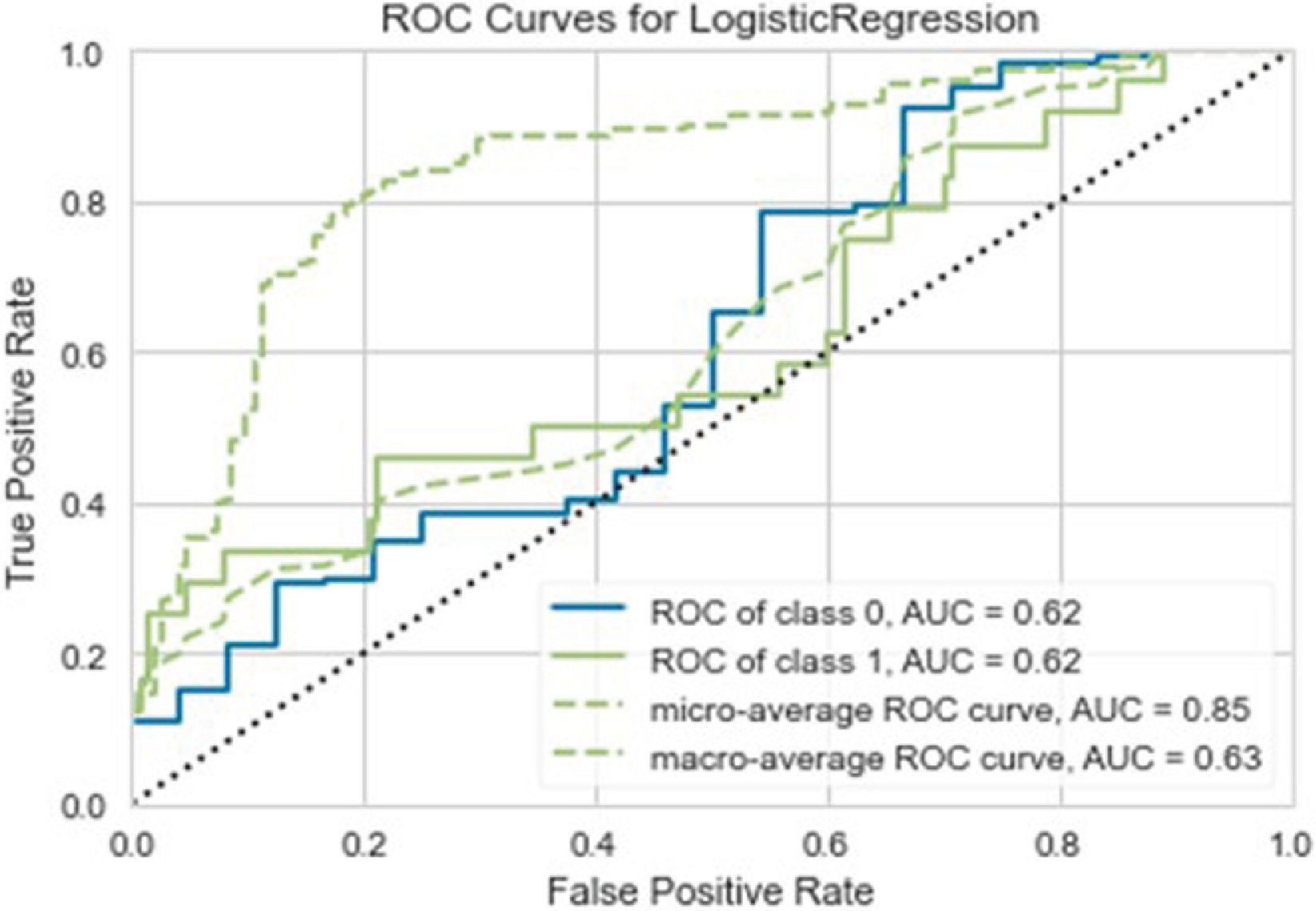
Although it was the model with the best AUC, as can be seen in Fig. 3, the ROC curve of the selected model is close to the diagonal, which means that linear regression has a very limited performance or hardly better than random prediction. This observation is best seen in the confusion matrix of the test data in Fig. 4, where 30 cases are misclassified, and 16 of the 24 cases of deceased patients are classified as alive. A sensitivity of 31%, an accuracy of 34%, and an AUC of .65 are achieved.
Finally, regarding the weight of the variables in the calculation of the model, Fig. 5 shows that no variable obtained predominant importance, the type of surgery being the most influential. Some specific implants were also important in the algorithmic calculation, such as the XXS Furlong® cemented hemiarthroplasty stem (JRI), or the 125° Gamma3® short intramedullary nail (Stryker). Moreover, the need for non-weightbearing after surgery affected the survival of these patients. In terms of medications, vitamin D, quetiapine, and bisoprolol had an influence on survival (positive for the former, negative for the latter two). It is striking that the variable ‘Other drugs’ appears among the 10 variables with the greatest weight. This variable is used to indicate that the patient has been prescribed a sclerostin inhibitor (Evenity® from Amgem). However, this anti-osteoporotic treatment was not indicated in our cohort. The fact that it appears in the list is due to the cases of patients in which this data appeared as a ‘missing value’ (.6%), to whom code 12 was imputed, which was of some importance for the algorithmic calculation of the linear regression, although the reason for this is unknown.
DiscussionIn the present paper we attempted to find a predictive model of death at 6 months after hip fracture in patients over 74 years by applying machine learning techniques with data from five different data sources. In a study on the Swedish National Hip Fracture Register (RIKSHOFT) a linear regression model was developed that predicted patients’ risk of death one year after hip fracture with a sensitivity of 75% (versus 31% in our work), an accuracy of 62% (versus 34% in our work), and an AUC of .74 (versus .65 in our work).7 It should be noted that the RIKSHOFT study included patients over 18 years of age (compared to 74 years in our study), and that the variable with the greatest weight in the algorithm was ‘metastatic carcinoma’, followed by ASA risk, sex, and age. In our case, several of the variables with the greatest weight in the algorithm have a clear pathophysiological reason, such as the need for non-weightbearing after surgery or the prescription of certain medications from which significant cognitive impairment (quetiapine) or congestive heart failure (bisoprolol) can be deduced. Others, such as the month of admission (January, February… December), or the use of certain specific implants, have no clear pathophysiological explanation.
Both the work undertaken on RIKSHOFT and ours have demonstrated the superiority of linear regression over other models which, a priori, could be assumed to be more powerful in generating predictions, such as the Gradient Boosting Classifier, as has been published by some authors in other disciplines of orthopaedic surgery and traumatology.8,9 It should be borne in mind that the more complex the model, the greater the amount of data required for training, which may explain the low performance of some of these algorithms, such as the Gradient Boosting Classifier or the Ada Boost Classifier.
Our cohort had a mortality rate of 19.2% at 6 months, similar to that recently published by other Spanish authors.10
Finally, although in general when working with various data sources, SQL databases are used, the NoSQL database MongoDB has shown great flexibility and performance, in addition to being easily integrated into Python code through the Pymongo module. Its use in conjunction with the different Python libraries makes it possible to solve the challenge posed by these databases in terms of reporting and analysis needs.11
ConclusionsDespite the significant volume of data used in training the models, we were not able to generate one that accurately predicts the vital status of patients over 74 years old 6 months after admission for hip fracture. It may require a larger number of patients and using more variables, which could be obtained from other sources (e.g., clinical analyses, personal history, or pre-admission treatments). We therefore believe that the method used for data extraction, transformation, and load, as well as for handling missing values using a NoSQL MongoDB database, and the Python modules Pandas, Pymongo, and Pycaret, can serve as a reference for future work.
Level of evidenceLevel of evidence iii.
Ethical considerationsThe Ethics Committee of the Hospital Universitario Galdakao-Usansolo approved this research. The authors complied with the relevant ethical standards for publication. The CEIC did not require informed consent from patients.
FundingFunded by the authors themselves.
Conflict of interestsThe authors have no conflict of interests to declare.