The "Cryptococcus neoformans species complex" is a denomination for the diverse yeasts that cause cryptococcosis, one of the most prevalent life-threatening mycoses10,28,33. Since the first descriptions of C. neoformans, many intraspecific differences have been described and a complicated sub-classification has been established within the species17,23,25. Further studies revealed several differences between the anamorphs of the two described varieties (C. neoformans var. neoformans and C. neoformans var. gattii) regarding ecology, epidemiology, pathogenicity, biochemistry and genetics5,6. Thanks to molecular biology tools, the existence of two species (Cryptococcus neoformans and Cryptococcus gattii) has been well established26. Each of them corresponds to two distinct set of serotypes. In the case of C. neoformans the serotypes are A and D, and two varieties are proposed for each of them (C. neoformans var. grubii and C. neoformans var. neoformans) but the existence of hybrid strains with an AD serotype is well known5. For C. gattii the serotypes are B and C. Nevertheless, some anomalous serotypes BD have been described. These strains could be considered as hybrids between the two species C. neoformans and C. gattii5.
Although serotyping was the first method used to study the epidemiology of cryptococcosis, the analysis of genomic patterns obtained after amplification of minisatellite/microsatellite sequences, intergenic spacer regions and/or the digestion of some specific genes improved the sensitivity and specificity of strain typing1,10,13,20,21,29,31. These molecular tools provide a new possibility for the epidemiological survey of Cryptococcus infections and ecological assessment of the source of the disease.
Cryptococcosis by C. neoformans is cosmopolite mostly in immunocompromised patients. C.gattii has more aggressive behaviour and causes infections more frequently in immunocompetent patients24. Until recently, this species has been considered to be restricted to warm areas (tropical and subtropical climates). These free-living yeasts can be isolated from soils, avian excreta and plant material1,6. However, the outbreak of cryptococcosis by C. gattii in the temperate climate of Vancouver Island (British Columbia, Canada)22 indicated a shift in the ecological niche of this species.
In the last decade, a number of DNA typing techniques have been used to study the epidemiology of C. neoformans. PCR fingerprinting has been used as the major typing technique for molecular epidemiological surveys of C. neoformans species complex12. In 1999 Meyer et al31 described eight major molecular types, VNI-IV for C. neoformans strains, and VGI-IV for C. gattii strains, which were proposed as epidemiological markers. The strains were typed using M13 PCR fingerprinting and orotidine monophosphate pyrophosphorylase (URA5) gene restriction fragment length polymorphism analysis (RFLP). In 2003 the same group carried out a study of more than 400 clinical and environmental isolates from Latin-American countries and Spain30.
Our laboratory leads the coordination of the Spanish cryptococcosis survey - within the European Conference of Medical Mycology Societies (ECMM) Cryptococcosis Working Group - which allowed us to type a large number of Spanish strains from environmental, human and animal samples8,9. Some of them were previously studied with pulsed field gel electrophoresis (PFGE) and their electrophoretic karyotypes showed a very high genomic variability19. At that time, there was no clear explanation for that genomic variability and no relationship with pathogenic aspects could be established. The possibility of studying the strains with newly proposed molecular markers encouraged the present work.
Three molecular techniques evaluating different molecular markers have been assayed for strain typing in this study. These were considered as low discriminative (5S rDNA RFLP analysis), moderately (URA5 RFLP analysis) and highly discriminative methods ([GACA]4 PCR fingerprinting)35. Therefore, each could provide a different applicability in epidemiological studies, i.e. [GACA]4 PCR fingerprinting is a good method for discrimination between highly related strains from the same origin. On the other hand, 5S rDNA RFLP analysis is a good discriminative tool to differentiate between C. neoformans and C. gatti species and varieties. The results obtained by URA5 RFLP analysis allowed classification of the strains in previously defined genotypes and comparison with other epidemiological studies.
Material and methods
Yeast isolates
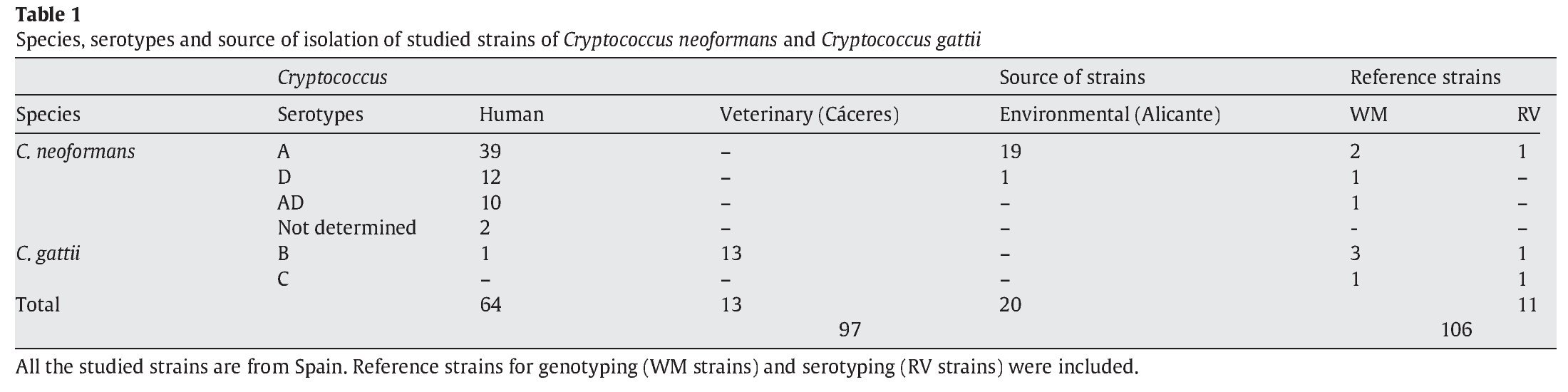
A total of 97 strains of C. neoformans species complex were analysed in this study. All of them were isolated in Spain: 20 from pigeon droppings, 64 from human clinical samples and 13 from veterinary samples (goats)2. In addition, 3 reference isolates provided by J.M. Torres (IMIM, Barcelona, Spain) and 8 from W. Meyer (University of Sydney, Australia), were used as controls for reproducibility of the techniques3,31. Detailed information for the isolates is provided in Table 1. As indicated in Figure 2, four pairs of strains from clinical samples were isolated from the same patient at different times during their disease follow-up.
Phenotype characterization
All of the isolates were identified as belonging to the C. neoformans species complex by standard methods: yeast and colony morphology, ability to grow at 30 and 37 °C on Sabouraud dextrose agar (SDA; Oxoid Ltd., Basingstoke, Hampshire, England), urease production (Urease Christensen), presence of capsule by India ink negative stain, pigment production on Staib medium, colorimetric sugar assimilation, actidione sensitivity and phenoloxidase production (Auxacolor Bio-Rad, France). Isolates were grown on Canavanine Glycine Bromothimol Blue (CGB) medium to discriminate between C. neoformans and C. gattii32.
Serotyping
The strains were serotyped by latex agglutination with the Crypto Check commercial kit (Iatron Laboratories, Tokyo, Japan) according to the manufacturer's instructions.
Genotyping
High molecular weight genomic DNA was extracted from cryptococcal cells by a modified method16 based on the standardized protocol described by White et al39. At least two independent DNA preparations were performed for each isolate. PCR reactions were carried out in a final volume of 50 ml. Each reaction contained 1× PCR buffer (75 mM Tris-HCl pH 9; 3 mM MgCl2; 50 mM KCl; 20 mM (NH)4SO4; 0.001% BSA; Biotools B&M Labs, S.A., Madrid, Spain), a final concentration of 200 µM of dNTPs, 1 U Taq DNA polymerase (Biotools B&M Labs, S.A., Madrid, Spain), 25 pmol of each indicated primer and 5 ml of extracted DNA. All PCR reactions were carried out in an Eppendorf Mastercycler gradient® (Eppendorf, Hamburg, Germany).
Amplification products from each PCR reaction were mixed with 20 ml of MilliQ water and separated by 2% (PCR fingerprinting) or 3% (URA5 RFLP products) agarose gel electrophoresis at 60 V for 2 or 3 h (PCR fingerprinting and RFLP analysis, respectively). Patterns were assigned visually by comparison with the molecular standard (Gene Ruler 100 pb DNA ladder Plus, MBI Fermentas, Vilnius, Lithuania). Images were captured by DigiDoc-It System v.1.1.27 (UPV, Cambridge, UK).
- 5S rDNA RFLP analysis: PCR amplification was carried out with primers 5S1 (5' GATCTACTGAGGCTAAGCCC 3') and 5S2 (5' AGACAAGCATATGACTACTG 3'). Conditions were adjusted as follows: 30 cycles; 95 °C 5 min initial denaturation, 1 min of denaturation at 95 °C, 2 min annealing at 52 °C, and 3 min extension at 72 °C, followed by a final extension cycle of 10 min at 72 °C11,14. Five microliters of PCR products were digested with 1 U of CfoI (ROCHE, Barcelona, Spain) for 2 h at 37 °C.
- URA5 RFLP analysis: PCR were performed with primers URA5 (5' ATGTCCTCCCAAGCCCTCGACTCCG 3') and SJ01 (5' TTAAGACCTCTGAACACCGTACTC 3'). Conditions were adjusted as follows: 35 cycles at 94 oC for 2 min initial denaturation, 45 s of denaturation at 94 °C, 1 min annealing at 61 °C and 2 min extension at 72 °C, followed by a final extension cycle for 10 min at 72 °C30. Fifteen microliters of PCR products were double digested with Sau96I (10 U/ml) and HhaI (20 U/ml) for 3 h at 37 °C.
- [GACA]4 PCR fingerprinting: the microsatellite-specific primer [GACA]4 (5' GACAGACAGACAGACA 3') was used as a single primer in the PCR reactions7,30. Amplification conditions were 40 cycles at 94 °C for 5 min initial denaturation, 30 s of denaturation at 94 °C, 45 s annealing at 43 °C and 30 s extension at 72 °C, followed by a final extension cycle for 10 min at 72 °C.
Data analysis
The NTSYS-pc version 1.8 (Applied Biostatistics, Inc.) was used to express results in a quantitative form. DNA bands obtained with each molecular method were defined manually. Similarity coefficients were estimated using the Jaccard algorithm and cluster analysis was performed by the Unweighted Pair Method with Arithmetic mean (UPGMA)34.
Reproducibility of the techniques
To assess intra-laboratory reproducibility of the methods of DNA extraction, PCR amplification and digestions were repeated three times with each strain. Inter-laboratory reproducibility of the methods was tested using reference strains. The eight molecular profiles described in the literature for URA5 RFLP analysis were obtained by including reference strains in the analysis. For [GACA]4 PCR fingerprinting, previous works have reported the high variability of the patterns7,30 and subsequently the inter-laboratory instability. Only intra-laboratory reproducibility could be assessed by obtaining the DNA fingerprinting for each strain at least three times.
Results
Phenotypes and serotype diversity of the isolates
After performing the different tests described in Materials and methods, it was determined that 83 (84.7%) of the isolates corresponded to C. neoformans and 14 (15.3%) to C. gattii. All studied isolates showed positive results for urease and phenoloxidase production, and growth at 37 °C. Carbohydrate assimilation showed 10 different patterns. Lactose was always negative and inositol was positive for 98%. The most frequent phenotypes were Auxacolor 75774 and 75775, with almost the same frequency of isolates, 37 (38.1%) and 34 (35.1%), respectively. All other auxonotypes presented a low (5-10 strains) or very low rate (< 5 strains).
The serotyping results showed that 58 isolates were serotype A (59.8%), which was the most frequent; 13 isolates (13.4%) were serotype D and 10 (10.3%) belonged to the AD hybrid. All the 14 C. gattii strains (14.4%) were serotype B. Two strains could not be serotyped (3.1%). Serotype C was absent in our study but reference strains were used to obtain and compare other possible molecular patterns (Table 1).
Genotype analysis of the isolates
Molecular profiles obtained by RFLP of the amplified genes IGS-5S and URA5 showed a number of different molecular patterns, which could be analysed manually by visual comparison of profiles. Data obtained after PCR fingerprinting with the microsatellite [GACA]4 showed a very high intra-species variability. For analyses, such high variability required the use of a specialized software that compared patterns and then clustered strains in dendrograms.
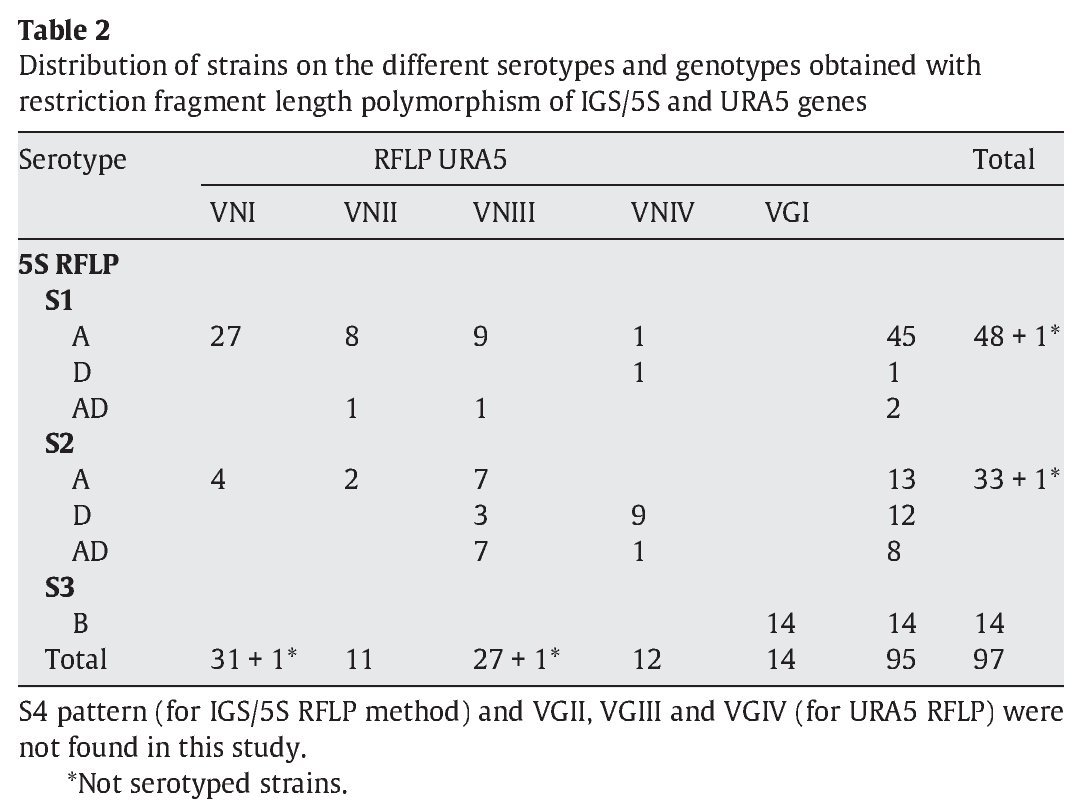
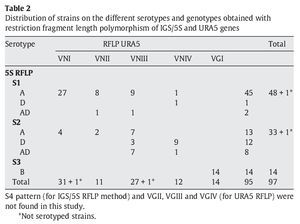
IGS RFLP (5S) analysis grouped the isolates in three major previously described genotypes (5S1-5S3). This molecular marker clearly differentiates the two species and varieties. For S1 and S2 types, 100% belonged to C. neoformans, and 100% of S3 types belonged to C. gattii isolates (VGI genotype). The S1 pattern was the most frequent among our isolates, which mainly corresponded to VNI/VNII C. neoformans var grubii strains (Table 2). The S4 pattern was not found in our study.
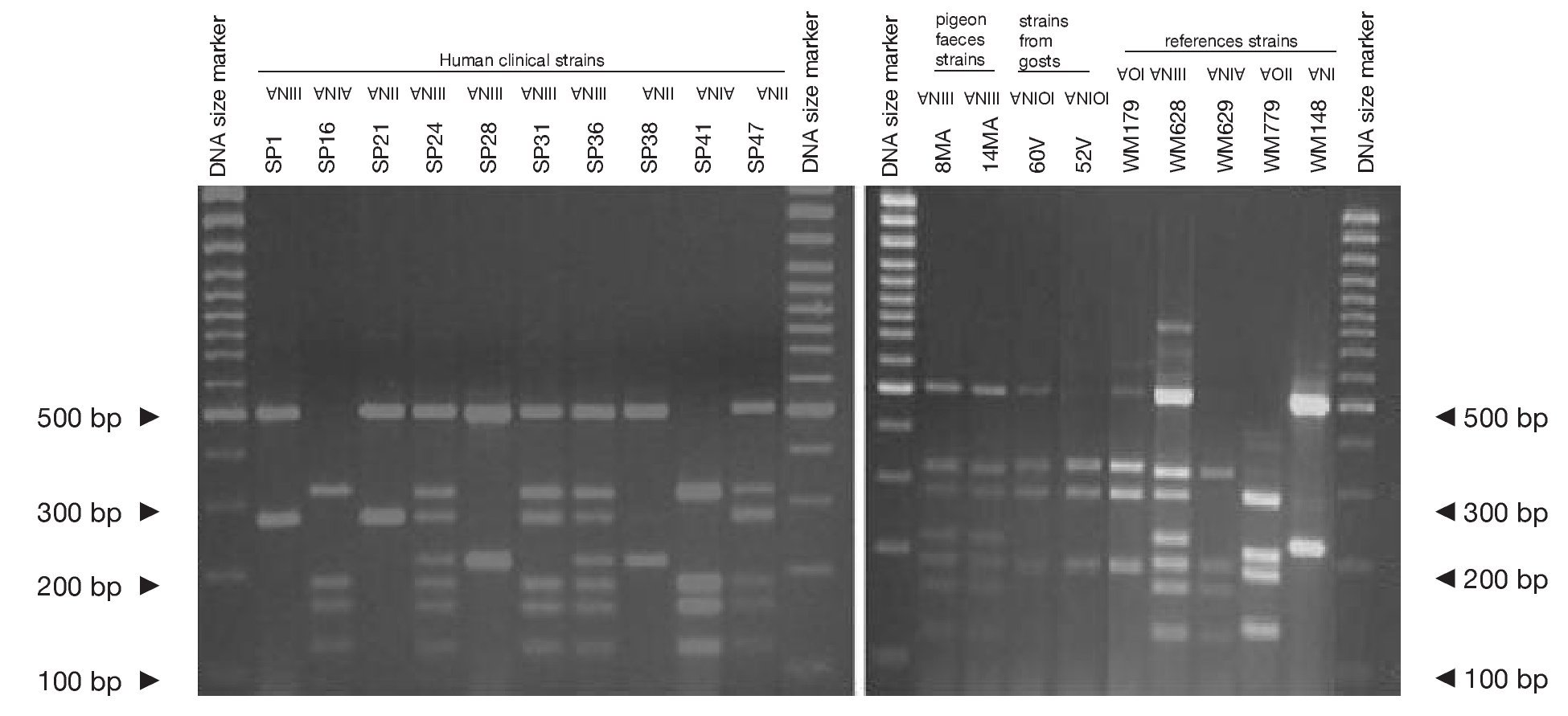
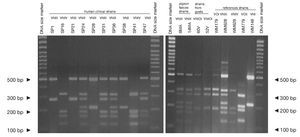
Concerning URA5 gene RFLP analysis, analyses of our isolates showed five of the eight patterns described previously30,31. Figure 1 shows a typical result of this kind of analysis. The profiles VNI and VNIII showed the highest frequency (33% and 28.9%, respectively). Profiles VGI (14.4%), VNIV (12.4%) and VNII (11.3%) had similar frequencies The virulent VGII genotype, which is related to some C. gattii B strains, was not found in this study22. VGIII and VGIV profiles were also absent among the studied isolates. All our serotype B strains belonged to the VGI molecular type (Table 2).
Figure 1. RFLP patterns obtained after double digestion of the URA5 gene. SP strains: isolates from human clinical samples; MA strains: isolates from pigeon faeces; V strains: isolates from goat tissues; WM strains: reference strains used to reproduce the molecular patterns VNI, II, III, IV, VGI and VGII, according to the molecular standards previously described (see Material and methods).
Reference strains included in the study showed identical profiles for URA5 gene RFLP (Fig. 1) as previously published30,31, providing us with appropriate tools for pattern identification and assessment of reproducibility.
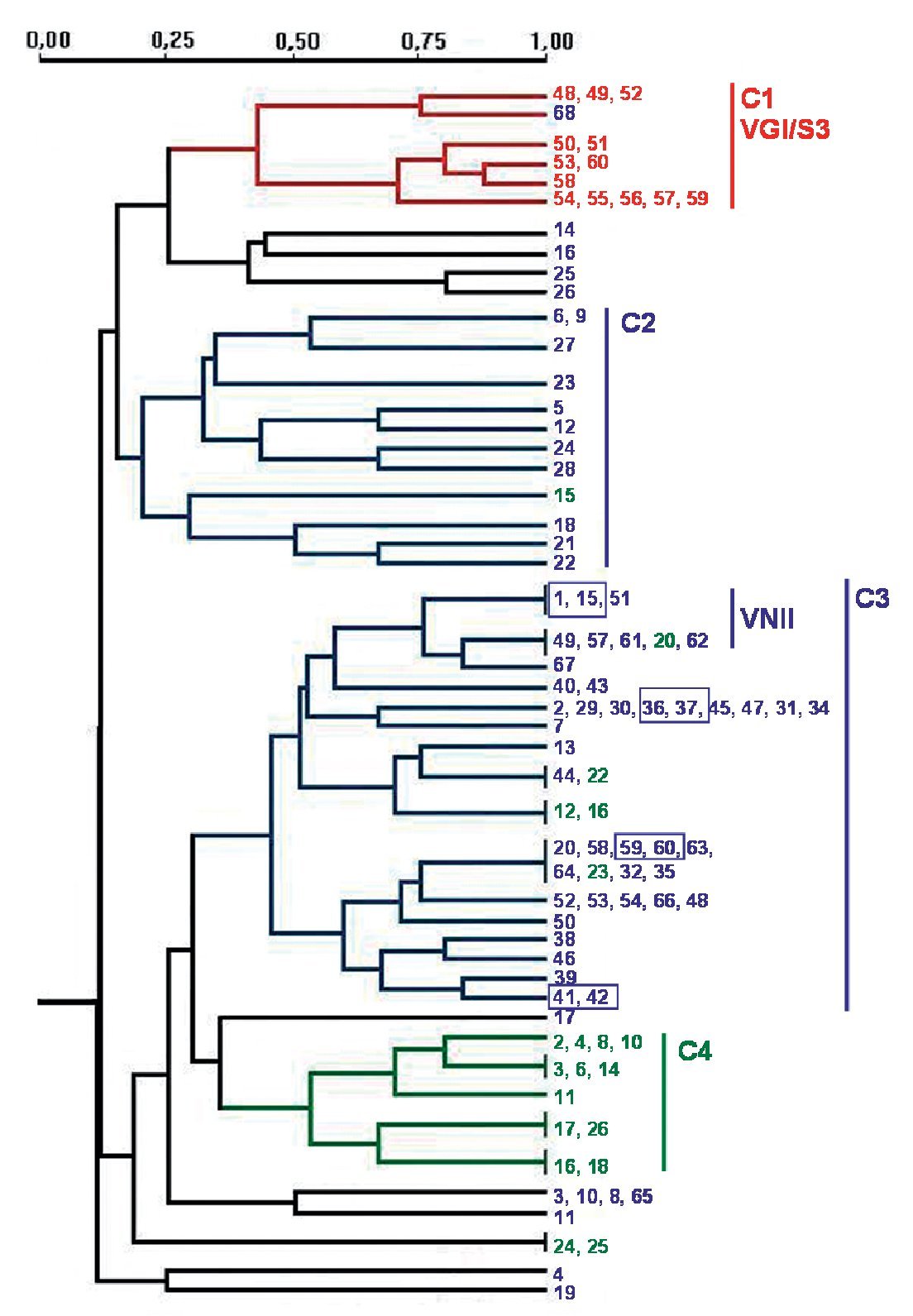
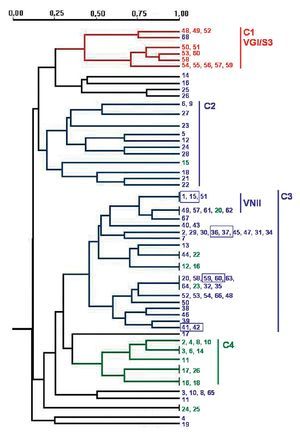
PCR fingerprinting with the microsatellite [GACA]4 showed 48 different profiles. Patterns were described taking into account each band that was constant for the same strain in successive DNA amplifications (repetitive analysis). This molecular marker showed the highest diversity. Results were analysed using the NTSYS software34, which clustered the strains on the basis of their similarity. The dendrogram in Figure 2 summarizes the results that were obtained. Strains were grouped in four main clusters, which show a good correlation with the different origin of the isolates. Cluster 1 groups the veterinary isolates, clusters 2 and 3 harbour 95.3% of the clinical isolates and cluster 4 consists of 60% of the environmental strains. The four pairs of strains isolated from the same patient always appear in the same branch in identical yeasts. Isolates of VGI and VNII genotypes clustered together in the dendrogram. Nevertheless, other genotypes (VNI, VNIII and VNIV) are randomly distributed in the groups of C. neoformans isolates (Fig. 2).
Figure 2. Dendrogram obtained with molecular profiles of all studied isolates after [GACA] amplification. Blue numbers correspond to strains obtained from human clinical samples (SP1-SP68). Green numbers: environmental strains from pigeon droppings (MA1-MA45); red numbers: strains from goat tissues (48V-60V). Blue boxes indicate strains obtained from a same patient. Main clusters identified by numbers from C1 to C4.
Geographical distribution of the isolates
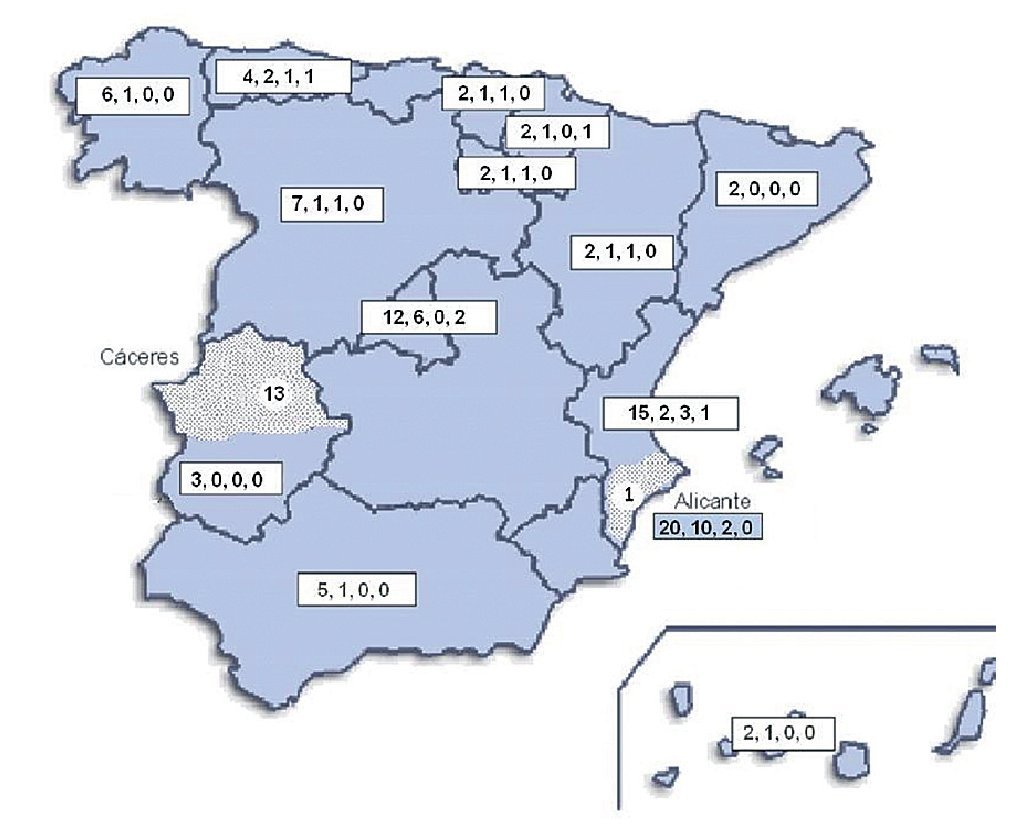
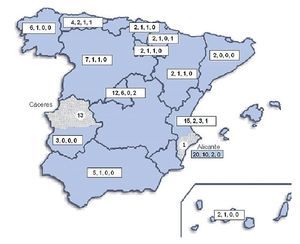
Geographical distribution of the strains in the map of Spain (Fig. 3) shows a random distribution of all the genotypes obtained from clinical samples. The URA5-VNIII profile, which would be considered especially important in our country, was detected in different parts of our geography. Regarding environmental strains, the high presence of this VNIII genotype among the isolates from pigeon droppings (50%) obtained in Alicante is remarkable. C. gattii was detected in two different regions of the country that were approximately 700 km apart. All the strains obtained from goats in Cáceres showed a high clonicity and had the same genotype with all the molecular markers used. The unique isolate of this species obtained from a human patient in Alicante also showed the same genotype.
Figure 3. Geographic distribution of the isolates studied. Numbers in the boxes indicate correlativelythe follows: first - total number of clinical isolates from that region of Spain; second - isolates showing URA5 VNI pattern; third - isolates showing URA5 VNIII pattern. Dotted areas correspond to the provinces of Alicante and Cáceres, where the environmental and veterinary strains were, respectively, isolated.
Discussion
During the last 10 years and from different parts of the world, a number of interesting reports have been published regarding the molecular typing of yeasts belonging to the C. neoformans species complex4,15,20,37,36. Besides the geographical differences reported for C. neoformans and C. gattii species, most of the results published showed that there is a predominant genotype for each of these two pathogenic species: VNI is the most prevalent profile among C. neoformans isolates all over the world and VGI within the C. gattii strains. In the present study, we also found VNI and VGI to be the most prevalent genotypes for each species in Spain. Nevertheless, the main finding in our work is, undoubtedly, the high presence of VNIII genotype of C. neoformans in our country.
The RFLP study of URA5 gene showed five of the eight previously described profiles (VNI-IV and VGI). The VGII, VGIII and VGIV profiles were absent among C. gattii serotype B strains and only VGI genotype was detected for this species. For C. neoformans isolates, VNI was the most prevalent type (33%) followed by VNIII (28.9%), which showed a very high prevalence when compared with other studies. These results agree with those obtained by Meyer et al30. on a limited number of Spanish isolates. In Meyer's study of Iberoamerican isolates, they found 42% of VNIII among Spanish strains. This genotype is absent in a number of studies from different parts of the world30,36 or had a very low presence in some others like the 3.6% and 1.5% in Argentina and Mexico15,30. Only in Chile, a significant presence of this profile had been described (15.8%).30 In our study, VNIII pattern had the highest presence among the environmental strains (50%) obtained from pigeon droppings, and also showed a very important presence within clinical isolates (28%).
This similarity between Chile and Spain is not restricted to the VNIII pattern. C. neformans genotype VNIV also had a remarkable presence in our study (12.4%), again similar to the rate found in Chile (26.3%) by Meyer et al30. This prevalence is higher than in other Latin-American countries, where the VNIV pattern was almost absent. VNIII pattern is mainly related to hybrid C. neoformans yeasts (AD serotypes) and VNIV to C. neoformans var. neoformans (D serotype)30. The high presence of D serotype in Europe is well assessed by some previous studies3,9,37,38. So the origin of the high prevalence of hybrid genotypes could be explained for the co-existence of the two haploid forms (A and D) in our geographical area. In this regard, it is important to remark that most of the VNIII strains in our study exhibited serotype A (59.3%), although 80% of the serotype AD isolates were also VNIII. This probably means that some hybrid forms of C. neoformans only show one serotype.
Amplification with microsatellite [GACA]4 revealed a highly discriminative power. Among the studied isolates, 48 different profiles were obtained. As in other studies, strains from a same species clustered together20,27 and we also found a good correlation between the PCR fingerprinting and the origin of the isolates. The dendrogram obtained with this method grouped the strains in four main clusters that correlate with the source of the isolates and, in some cases, with the molecular types (Fig. 2). The first cluster - C1 - harbours all VGI/S3 genotypes, which also correspond to all the C. gattii isolates studied. Therefore, all strains from veterinary origin were grouped here with the unique C. gattii of human origin. The low number of strains and the common origin of the veterinary ones (from a cryptococcosis outbreak among goats) make it difficult to assess that the [GACA]4 profile is specific to this origin, species or genotype. For C. neoformans strains, two big clusters grouped 95.3% of the clinical isolates and a third one grouped 60% of the environmental isolates (Fig. 2). It is also remarkable that 72.7% of the VNII isolates are grouped in the same branch inside the second clinical cluster - C3 - while other C. neoformans genotypes (VNI, VNIII and VNIV) appeared randomly distributed among the three clusters.
The specific case of the strains obtained from the same patient at different times during the follow-up of the disease showed that they were consistently grouped together in the same branch as identical strains. Other studies with [GACA]4 and [GTG]5 as molecular markers also proved that strains from the same patient, but from different parts of the body, always showed the same molecular type20,31. From these results, we can conclude that the diverse patterns obtained with [GACA]4 fingerprinting are stable (intralaboratoy) and thus, could be a useful method for either the follow-up of a strain or the differentiation between relapse and reinfection in patients who suffer from repetitive episodes of cryptococcosis.
Molecular patterns obtained by digestion of 5S-IGS amplified fragments showed three different profiles within the studied strains. These "S types" can be easily related with species, varieties and serotypes. In fact, serotype D and AD strains of C. neoformans belonged to the S2 profile at a significantly high rate (92.3% and 80%, respectively) and showed VNIII and VNIV genotypes (Table 2). Serotype A strains were distributed in S1 (77.6%) and S2 (22.4%) profiles. S1 type was the most prevalent and harboured most of C. neoformans VNI and VNII (C. neoformans var. grubii) isolates included in the study. Serotype B isolates (C. gattii, VGI) corresponded 100% to the S3 type. This method has been scarcely used by other authors11,14. So the results are somehow independent and difficult to compare. Therefore, inter-laboratory reproducibility could not be evaluated.
Nowadays, the source of most Cryptococcus infections is not very clear, and some epidemiological events, like the Vancouver outbreak, ought to make us think about the significant lack of knowledge we have about their route from the environment to human or animal patients, the worldwide spread of the species or genotypes of the yeast, and the different virulences they could display. The excellent work carried out with the Vancouver outbreak strains is a good example to reinforce the study of Cryptococcus typing22. The study based on some genotyping methods like URA5 RFLP, AFLP analysis and Multilocus Sequence Typing (MLST) showed a special situation in the Vancouver outbreak with a high spread of two subtypes of VGII molecular type18. From that moment on, a relationship between high virulence and genotype VGII could be established within C. gattii B serotype and some hypothesis of the probable spread from their origin was proposed.
The results shown here could indicate the importance of local environmental factors for strain distribution of this pathogen in a limited geographical area. We can confirm the high frequency of the VNIII profile previously described by Meyer and his co-workers in their Iberoamerican's strain study. This fact, together with the still unexplained goat's cryptococcosis outbreak that occurred in Cáceres some years ago, makes our country different from other European and Mediterranean areas regarding the epidemiology and ecology of Cryptococcus species complex. The similarity between Spain and Chile in the prevalence of some C. neoformans var. neoformans and hybrid strains could be an interesting feature to explain the spread of this yeast around the world.
Acknowledgements
The authors wish to acknowledge the financial support from Fundación Navarro-Tripodi of Alicante and Generalitat Valenciana (GV-00-045/3), Wieland Meyer and Josep María Torres for providing the reference strains, members of the Spanish and the ECMM Cryptococcosis working groups for providing clinical strains and María Abad for the English review of the manuscript.
* Corresponding author.
E-mail address: colom@umh.es (M.F. Colom-Valiente).
ARTICLE INFO
Article history:
Received August 19, 2008
Accepted December 2, 2008