Aspergillus fumigatus is a ubiquitous opportunistic pathogen. This fungus can acquire resistance to azole antifungals due to different mutations in the cyp51A gene. Azole resistance has been observed in several continents and appears to be a globally distributed phenomenon. Specific mutations in cyp51A that lead to azole resistance, such as the TR34/L98H modification, have been reported.
AimsTo evaluate the azole resistance in clinically isolated A. fumigatus strains.
MethodsAs a result of our passive surveillance strategy, a total of 23 A. fumigatus isolates from clinical origins were identified through a phylogenetic analysis using the ITS region and β-tubulin gene fragments, and typed with the CSP microsatellite. Azole susceptibility profiles were performed by disk diffusion and microdilution broth methodologies according to CLSI guidelines.
ResultsHere we describe, for the first time, the detection of azole-resistant A. fumigatus isolates from clinical origins in Chile with mutations in the cyp51A gene. In addition to the TR34/L98H mutation, one isolate exhibited an F46Y/M172V/E427K-type mutation. Furthermore, microsatellite typing based on cell surface protein (CSP) was performed, showing the t02 (TR34/L98H), t15 (F46Y/M172V/E427K) and t01 (susceptible clinical isolates) genotypes.
ConclusionsOur study demonstrates the presence of mutations related to azole resistance in A. fumigatus strains isolated from clinical samples in Chile. In order to obtain information that may help to tackle the spread of antifungal resistance among A. fumigatus populations, and to ensure the efficacy of future treatments against aspergillosis, a further research is necessary.
Aspergillus fumigatus es un patógeno oportunista ubicuo. Este hongo puede adquirir resistencia a los antifúngicos azólicos gracias a diferentes mutaciones en el gen cyp51A. La resistencia a los azoles se ha observado en varios continentes y parece ser un fenómeno de distribución mundial. Se han descrito mutaciones específicas en cyp51A para adquirir resistencia a los azoles, como la modificación TR34/L98H.
ObjetivosEvaluar la resistencia a los azoles en aislamientos de A. fumigatus obtenidos de muestras clínicas.
MétodosGracias a la vigilancia pasiva se identificó un total de 23 aislamientos de A. fumigatus de origen clínico a través de un análisis filogenético mediante el estudio de la región ITS y de fragmentos del gen de la β-tubulina, que se tipificaron con el microsatélite CSP. Los perfiles de sensibilidad a los azoles se realizaron mediante los métodos de difusión en disco y microdilución.
ResultadosEn el presente trabajo describimos, por primera vez en Chile, la detección de aislamientos clínicos de A. fumigatus en este país resistentes a azoles, con mutaciones en el gen cyp51A. Además de la mutación TR34/L98H, un aislamiento presentó una mutación de tipo F46Y/M172V/E427K. Se realizó la tipificación por microsatélites basada en la proteína de superficie celular, que mostró los genotipos t02 (TR34/L98H), t15 (F46Y/M172V/E427K) y t01 (cepas sensibles).
ConclusionesNuestro estudio demuestra la presencia de mutaciones relacionadas con la resistencia a los azoles en cepas de A. fumigatus aisladas de muestras clínicas en Chile. Para obtener información que pueda ayudar a atajar la propagación de la resistencia a los antifúngicos entre las poblaciones de A. fumigatus, y garantizar así la eficacia de futuros tratamientos contra la aspergilosis, es necesario seguir investigando.
In recent years, the number of pathogenic fungi and the incidence of mycoses have increased considerably. Among the causes that may explain this situation, the increase in the population at risk of contracting an invasive fungal disease (IFD), i.e. immunocompromised patients, those with oncohematological problems, or those with prolonged corticosteroid treatment must be mentioned.39 Another cause is the improvement in both the diagnosis of mycoses and the identification of the etiological agents. Nevertheless, IFDs still have a high morbidity and mortality rate, which can reach 90% in some groups of patients.4,7,35,36
Among the different fungi capable of producing IFD, we can find two large groups of etiological agents: yeasts, with Candida albicans being the most representative, and filamentous fungi, with Aspergillus fumigatus as the most isolated microorganism.
The genus Aspergillus is a diverse group of fungi comprising many species capable of producing a wide variety of pathologies. Classically, the identification of the different species has been based on the phenotypic characterization of the isolates. However, due to the improvement and development of new molecular tools in the last decades, these advanced methodologies are fundamental in the identification of the Aspergillus species. Currently, this genus contains about 446 species. However, only some of them are pathogenic for humans, being the species belonging to the sections Fumigati, Flavi, Nidulantes, Nigri, Terrei and Usti the most commonly isolated from clinical cases.29
We daily inhale a large number of Aspergillus spores, most of which get trapped in the epithelium of the lower respiratory tract and are eliminated by the ciliary clearance mechanism. Only the smaller spores (between 2 and 5μm) are able to reach the pulmonary alveoli, where they will be removed by the alveolar macrophages, responsible for eliminating the conidia, and the neutrophils, responsible for destroying the few hyphae produced from the germination of the spores.27 However, in immunocompromised patients, the presence of the spores can lead to colonization of the host, enabling the development of IFD.9,33
Diseases caused by species of the genus Aspergillus are generally referred to as aspergillosis. This genus is responsible for more than 200,000 cases of invasive aspergillosis (IA) per year. Besides, this genus causes more than 1.2 million additional cases of chronic pulmonary aspergillosis (CPA) and about 4.8 million cases of allergic bronchopulmonary aspergillosis (ABPA).17,18 In Chile, due to the absence of official data about IFD, cases of IA, CPA and ABPA are estimated to occur in 1.7/100,000; 6.9/100,000 and 97.9/100,000, respectively.2
One of the threats related to aspergillosis is the resistance to antifungals, mainly azoles, which are the treatment of choice in these mycoses. Such resistance seems to be caused by various mutations in the Cyp51A gene, which codes for 14-α-demethylase, the target enzyme of azole drugs,34,45,46 and which would be related to high rates of therapeutic failure. The first reports of this phenomenon date back to the 1990s,16 being an emerging problem widely reported in many countries around the world.
The present work reports the isolation for the first time in Chile of azole resistant strains of A. fumigatus. We also prove the mutations in the Cyp51A gene.
Material and methodsFungal isolates and morphological identificationTwenty-three clinical isolates obtained from March 2017 to March 2021, classified as Aspergillus spp., were received to perform the species identification. The isolates were obtained by culturing bronchoalveolar lavage samples from patients diagnosed with aspergillosis. Firstly, the isolates were identified by a phenotypical approach, evaluating their macro- and microscopic features and their ability to grow in the dark at 10°C, 25°C, 37±1°C, and 50°C on potato dextrose agar (PDA).
Molecular analysisAccording to the manufacturer's instructions, the DNA was extracted using an E.Z.N.A.® Fungal DNA Mini Kit (Omega Biotek Store, USA). The universal fungal primers ITS1 and ITS4 for ITS1-5.8S-ITS2 region,47 and Bt2a and Bt2b24 for a partial β-tubulin gene sequence, were used. A 30μl mixture containing 17μl of BioMixTM Red (New England Biolabs), 10pM primers, and 10ng DNA was subjected to a DNA amplification technique, following a protocol of initial denaturation at 94°C for 5min, 35 cycles of denaturation at 94°C for 1min, annealing at 52°C for 1min for the ITS region, and 55°C for β-tubulin; extension at 72°C for 2min, and final extension at 72°C for 7min. The PCR product was checked on 2% agarose gel. Purification was performed following manufacturer guidelines with FavorPrep™ Gel/PCR purification Mini Kit (Favorgene, Taiwan). Sequencing was carried out in Macrogen (Macrogen, Korea). An NCBI Blastn search for each locus region was performed.1 Sequences from type and reference strains were retrieved from the GenBank database. The sequences were aligned in MUSCLE software20 followed by manual adjustments with a text editor. ITS sequences were used to identify isolates to the Aspergillus-section level, and partial β-tubulin sequences were used to identify to the species level.
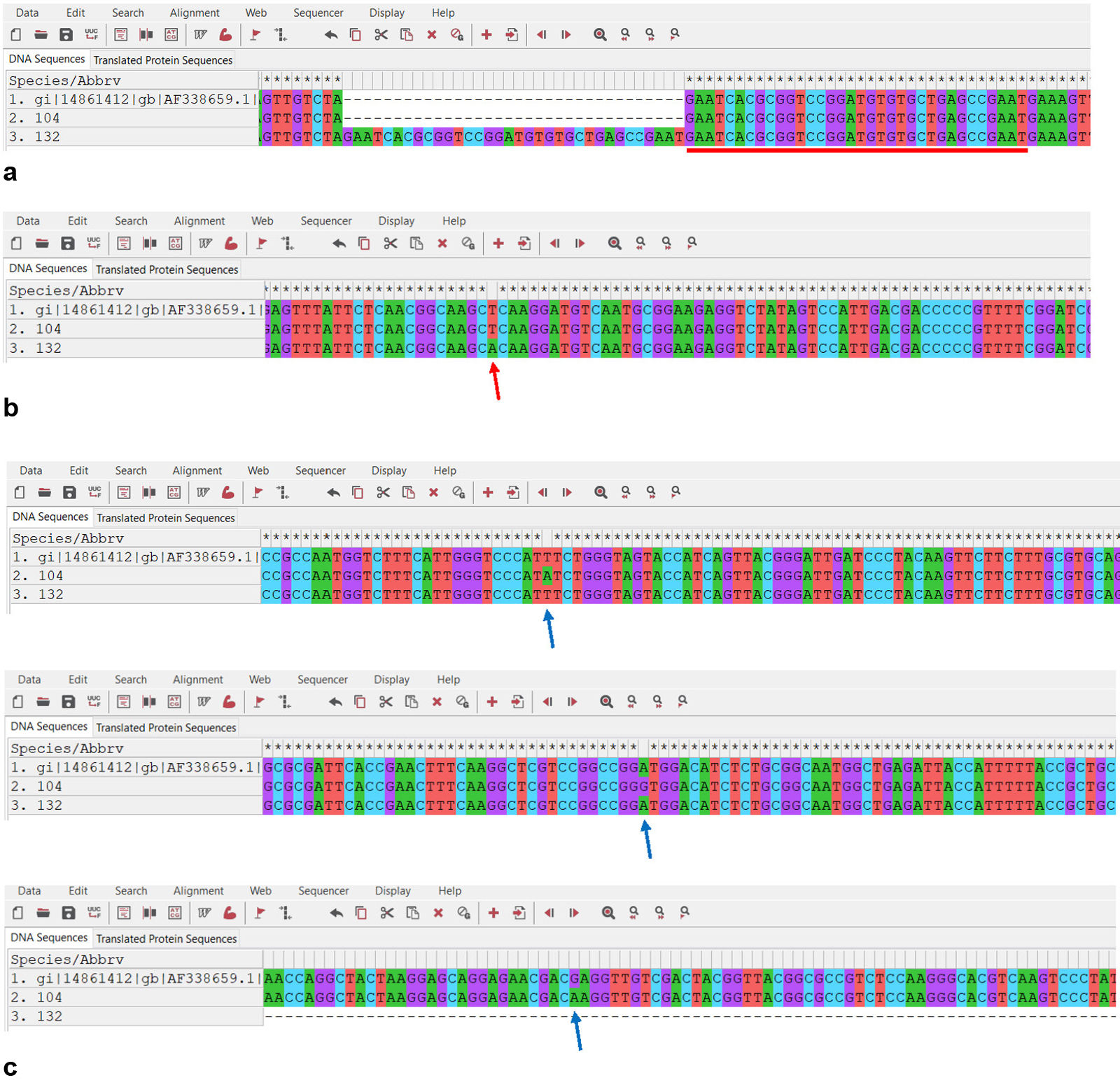
The entire sequences of the Cyp51A gene and promoter regions were amplified as previously described42 in any isolate that exhibited azole resistance with the purpose of finding mutations that might explain that condition. Sequencing and analyses were carried out as formerly mentioned. The DNA sequences were compared with the wild-type susceptible A. fumigatus reference strain (GenBank AF338659).
Finally, the cell surface protein (CSP) encoding gene was partially sequenced8 in all the isolates to perform the CSP typing according to the nomenclature reported by Klaassen et al.32
Antifungal analysisThe in vitro susceptibility to itraconazole (ITZ), posaconazole (PCZ) and voriconazole (VRZ) was evaluated by the broth microdilution method performed according to the CLSI document M38M51S.13 The antifungal concentration of ITZ, PCZ and VRZ ranged from 0.0156 to 8μg/mL. The inoculated plates were incubated at 35°C and observed after 24h. The lack of visual growth in each well was defined as the MIC value. Due to the absence of breakpoints for filamentous fungi in the CLSI method (except for voriconazole and A. fumigatus), the epidemiological cut-off values (ECVs) were used (1mg/L for ITZ and VRZ, 0.5mg/L for PCZ).14,10,22 In order to confirm the obtained results, the susceptibility test was performed twice. Candida parapsilosis ATCC 22019, A. fumigatus ATCC MYA-3626 and Hamigera insecticola (ex. Paecilomyces variotii) ATCC MYA-3630 were used as controls.
In addition, an antifungal susceptibility test by the disk diffusion method was performed according to the CLSI M38M51S document.13 Isolates were subcultured on PDA at 25°C for 5–7 days before testing. Then, the surface of a Mueller Hinton agar (MHA) plate dish was inoculated with a sterile cotton swab with the undiluted mold stock inoculum suspension. Antifungal disks (Rosco Diagnostica, Denmark) including ITZ (10μg), PCZ (5μg), and VRZ (1μg) were applied onto the surface of the inoculated media. The plates were incubated at 35°C and read after 24–48h. Zone diameters were interpreted according to CLSI M38M51S. A slight trailing or hyphal element extending into the inhibition zone was ignored.
ResultsThe twenty-three strains analyzed in the present study were identified as Aspergillus section Fumigati based on their morphological and physiological characteristics: blue-green colored velutinous colonies, and abundant sporulation on PDA. The isolates showed subclavate vesicles (13–26μm), uniseriate columnar heads and flexuous conidiophore. No strain grew at 10°C, but did it at 50°C on PDA, as expected according to the literature.28,40 The analysis of the nucleotide sequences of the β-tubulin gene showed that two strains had a percent identity equal to or higher than 99% with those sequences of A. fumigatussensu stricto available in the GenBank/NCBI database (99.6% sequence of the strain >EF669851.1 NRRL 5587).
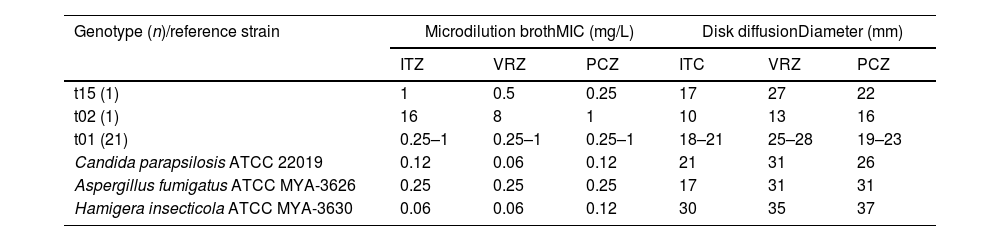
Antifungal susceptibilityTable 1 shows the antifungal susceptibility profiles obtained by means of the microdilution method and the disk diffusion method. Voriconazole and posaconazole showed the highest activity. One isolate (ChFC 132) was resistant to voriconazole with a MIC of 8μg/mL. Also, and due to a lack of clinical breakpoints, ChFC 132 was considered as non-wild type (NWT) based on the proposed ECV, exhibiting MIC values of 16μg/mL for ITZ and 1μg/mL for PCZ. The remaining 22 isolates showed MIC values of 1μg/mL, 0.5μg/mL and 0.25μg/mL for ITZ, VRZ and PCZ, respectively. Based on these results, these isolates were considered susceptible to VRZ and wild-type for ITZ and PCZ.
Antifungal susceptibility profiles obtained with the microdilution broth method and the disk diffusion method.
| Genotype (n)/reference strain | Microdilution brothMIC (mg/L) | Disk diffusionDiameter (mm) | ||||
|---|---|---|---|---|---|---|
| ITZ | VRZ | PCZ | ITC | VRZ | PCZ | |
| t15 (1) | 1 | 0.5 | 0.25 | 17 | 27 | 22 |
| t02 (1) | 16 | 8 | 1 | 10 | 13 | 16 |
| t01 (21) | 0.25–1 | 0.25–1 | 0.25–1 | 18–21 | 25–28 | 19–23 |
| Candida parapsilosis ATCC 22019 | 0.12 | 0.06 | 0.12 | 21 | 31 | 26 |
| Aspergillus fumigatus ATCC MYA-3626 | 0.25 | 0.25 | 0.25 | 17 | 31 | 31 |
| Hamigera insecticola ATCC MYA-3630 | 0.06 | 0.06 | 0.12 | 30 | 35 | 37 |
ITZ: itraconazole; VRZ: voriconazole; PCZ: posaconazole; MIC: minimal inhibitory concentration.
The results obtained with the disk diffusion method were consistent with those observed with the microdilution broth methodology. In brief, isolate ChFC 132 showed an inhibition zone ≤17mm for all triazoles tested. According to these values, ChFC 132 was considered NWT. In the remaining 22 strains, the inhibition halos observed were greater than 17mm for VRZ, PCZ and ITZ, being considered as susceptible to all the antifungals tested. However, one of these strains (ChFC 104) exhibited inhibition halos ≥17mm for VRZ and PCZ, but ≤17mm for ITZ. Interestingly, in the halo around the ITZ disk, a trailing zone was observed, which is why it was decided to sequence the CYP51A gene in order to elucidate putative mutations.
Cyp51A gene sequencing and CSP typingSequencing of the Cyp51A gene and its promoter showed the presence of the 34 bp tandem repeat (TR34) and L98H mutation in the strain ChFC 132. In addition, this analysis revealed that the second isolate (ChFC 104) harboured the F46Y/M172V/E427K mutations (Fig. 1). In the remaining isolates, no mutations were observed.
The characterization of the CSP showed that our strains corresponded to types previously described in the literature. The strain ChFC 132 belongs to type t02, while the ChFC 104 was identified as type t15. The other 21 strains were classified into type t01.
DiscussionInvasive aspergillosis is a very important challenge in medical mycology. An early diagnosis may be difficult to achieve, and the treatment of the disease is complex, so antifungal agents are used at the slightest suspicion of infection. In this context, the development of secondary resistance to commonly used antifungals gets facilitated. This fact is becoming a problem that is aggravated by the existence of cross-resistance between clinical triazoles and other azoles of the same family used as antifungals in agriculture.23
Likewise, azole resistance is now being reported in countries worldwide.5,19,44 In Latin America, several countries have reported the presence of azole resistance in A. fumigatus, such as Colombia,3 Argentina30 and Peru,11 among others. To our knowledge, this study describes for the first time in Chile the presence of azole resistance in A. fumigatus. Our two isolates with the mutations already mentioned were recovered from the bronchoalveolar lavage of two patients with aspergillosis. The samples were sent to our laboratory in order to check the presence of fungal organisms. Due to our passive surveillance strategy, we performed a disk diffusion test to evaluate their susceptibility patterns to some azoles. Based on the presence of halos ≤17mm (ChFC 132), as well as the presence of what appeared to be trailing (ChFC 104), the isolates were subjected to the broth microdilution method, and the sequencing of the Cyp51A gene. The first strain was harbouring the TR34/L98H mutations. These substitutions and tandem repeat sequences in the promoter region are some of the most commonly reported azole-resistance mutations.37,43 They were first described in Dutch A. fumigatus isolates, but are now spread worldwide19 due to the extensive use of azole fungicides in agriculture and animals. These mutations have been related to high MIC values to azoles in several global studies, especially to itraconazole and voriconazole. Our results show MICs over 1mg/L for VRZ and ITZ, and 0.5mg/L for PCZ, allowing us to classify this isolate as resistant for voriconazole, and NWT for itraconazole/posaconazole, respectively. This finding agrees with other reports demonstrating the pan-azole resistance that confers the presence of TR34/L98H.25,48
The F46Y/M172V/E427K substitutions were observed in the second isolate. Curiously, these mutations had been identified in both azole-susceptible and resistant A. fumigatus isolates.21,31 The F46Y/M172V/E427K mutations seem to be located in a protein region that does not interact with azole compounds or, at least, does not affect the structure of the protein.42 The above suggests that the mutations found might not be the molecular mechanism related to the observed resistance to azoles. In the same way, other situations, such as mutations of the HapE or Hmg1 gene, or efflux pumps, among others, could explain azole resistance in some isolates.12,26,38
In our study, three previously observed genotypes were identified. Most of the clinical strains analyzed in the present study were susceptible to all the antifungal assayed. Moreover, they were identified as the t01 genotype. Worldwide, the t01 genotype has been mostly associated with susceptible strains; however, resistant isolates have also been reported within that genotype. It seems that both susceptible and resistant isolates can be found in all genotypes. In fact, in a study carried out in the United Kingdom, no susceptible isolates were associated with the t01 genotype but with genotypes t03, t04A, t05, and t08.41 With respect to the other genotypes analyzed, isolate ChFC 132 was linked to t02 genotype, commonly associated with the TR34/L98H mutations.6 Likewise, the strain ChFC 104 was characterized as t15, a genotype reported in Europe and Australia. Interestingly, strains belonging to genotype t15 harboring F46Y/M172V/E427K mutations seem to be susceptible to all azoles.15 As we can observe, these genotypes have been recovered in several countries around the world, exhibiting a low genetic variation compared with wild-type strains. The detection of identical genotypes in distant countries suggests the clonal expansion, presumably airborne dispersal.41
ConclusionsWe report the finding of two clinical strains from Chile with mutations in the Cyp51A gene. The results obtained with both the disk diffusion method and the microdilution broth methodology were consistent. This demonstrates the usefulness and advantages of disk diffusion: its low cost and easy implementation in low complexity laboratories are the most remarkable features. Likewise, this methodology seems to predict azole resistance efficiently. Future studies involving more clinical and environmental strains are necessary to elucidate Chile's accurate picture of azole resistance.
Ethical approvalNot applicable.
FundingThis research received no external funding.
Authors’ contributionsConceptualization, E.A.D; Methodology, E.A.D., N.C., J.M.; Investigation, E.A.D., N.C., J.M.; Writing – original draft preparation, E.A.D., N.C.; Writing – review and editing, E.A.D. All authors have read and agreed to the published version of the manuscript.
Consent to participateNot applicable.
Consent for publicationNot applicable.
Competing interestsThe authors declare no conflict of interest.
Availability of data and materialsThe data that support the findings of this study are available from the corresponding author upon reasonable request.