Pneumocystis carinii is an opportunistic fungal pathogen that may cause pneumonia and lead to pulmonary fibrosis.
AimsThis study attempted to investigate the role of P. carinii infection-related genes in regulating lung fibrosis in mice.
MethodsA screening of P. carinii infection-related differential mRNAs was performed using the GEO database, followed by protein–protein interaction (PPI) network construction using the STRING website in order to obtain P. carinii infection-related key genes. The development of a mouse model with gene aberrant expression was achieved by utilizing mice carrying the Cre-LoxP recombinase system. Dexamethasone was employed to induce tracheal infection in order to develop a model of pulmonary fibrosis, and the magnitude of lung injury was assessed by performing hematoxylin–eosin (H&E) staining and Masson staining. Lung coefficient and hydroxyproline level were assessed on sections of lung tissue as well. Finally, the magnitude of lung fibrosis and inflammation in mice was determined based on immunofluorescence and on the expression of genes associated with lung fibrosis and inflammation.
ResultsFn1 was found by PPI with the highest connectivity in the PPI network associated with immunity and inflammation. Besides, Fn1 was significantly highly expressed in P. carinii-infected mice samples. The P carinii pneumonia (PCP)+Fn1fl/fl group had significantly higher lung coefficients, hydroxyproline levels and TNF-α, IL-6, IL-1β, IL-8 and NLRP3 expression levels, and significantly lower IL-10 expression levels. The results found in PCP+SPC-Cre:Fn1fl/fl group were the opposite. The results of the pulmonary fibrosis level study showed that the PCP+Fn1fl/fl group had the most intense H&E and Masson staining, and significantly higher expression levels of Col1A2, Col3A1 and α-SMA, which were lower in the PCP+SPC-Cre:Fn1fl/fl group.
ConclusionsP. carinii infection may promote the upregulation of Fn1, which causes pulmonary fibrosis with an inflammatory response.
Pneumocystis carinii es un patógeno fúngico oportunista que puede causar neumonía y provocar fibrosis pulmonar.
ObjetivosEste estudio trató de investigar el papel de los genes relacionados con la infección por P. carinii en la regulación de la fibrosis pulmonar en ratones.
MétodosSe realizó un cribado de moléculas de ARNm diferenciales relacionadas con la infección por P. carinii por medio de la base de datos GEO, seguido de la construcción de redes de interacción proteína-proteína (PPI) en el sitio web STRING con el fin de obtener genes clave relacionados con la infección por P. carinii. Se desarrolló un modelo de fibrosis pulmonar y se evaluó la magnitud de la lesión pulmonar mediante tinción con hematoxilina-eosina (H&E) y tinción de Masson. También se evaluaron el coeficiente pulmonar y el nivel de hidroxiprolina en secciones de tejido pulmonar. Por último, se determinó la magnitud de la fibrosis y la inflamación pulmonares en ratones mediante inmunofluorescencia y la expresión de genes asociados a la fibrosis y la inflamación pulmonares.
ResultadosFn1 se encontró por PPI con la mayor conectividad en la red PPI asociada con la inmunidad y la inflamación. Además, Fn1 se expresó significativamente en muestras de ratones infectados por P. carinii. El grupo con neumonía por P. carinii (PCP)+Fn1fl/fl presentó coeficientes pulmonares, niveles de hidroxiprolina y niveles de expresión de TNF-α, IL-6, IL-1β, IL-8 y NLRP3 significativamente más altos, y niveles de expresión de IL-10 significativamente más bajos. Los resultados encontrados en el grupo PCP+SPC-Cre:Fn1fl/fl fueron los opuestos. Los resultados del estudio del nivel de fibrosis pulmonar mostraron que el grupo PCP+Fn1fl/fl presentaba las tinciones de H&E y Masson más intensas, y niveles de expresión de Col1A2, Col3A1 y α-SMA significativamente superiores, que eran inferiores en el grupo PCP+SPC-Cre:Fn1fl/fl.
ConclusionesLa infección por P. carinii puede promover la expresión de Fn1, lo que causa fibrosis pulmonar con respuesta inflamatoria.
Pneumocystis carinii, by Delanoë and Delanoë, an opportunistic fungal pathogen, was found first in the lung of infected mice.33 After several changes in nomenclature, Pneumocystis jirovecii was the name proposed for the species causing pneumonia in human beings.29P. carinii is transmitted among animals through air, as animal experiments suggest. P. carinii pneumonia (PCP) occurs in animals carrying P. carinii after being immunosuppressed (reactivation of latent infection) and in immunocompromised animals after exposure to P. carinii.10Pneumocystis infection may be fatal in immunocompromised patients.7 As the most common infection among patients with advanced human immunodeficiency virus (HIV) infection, PCP significantly increases the mortality of VIH-AIDS people.16 Pulmonary fibrosis is one of the sequelae of Pneumocystis infection.37 As a chronic lung injury, pulmonary fibrosis results in the gradual destruction of lung tissue structure and the formation of fibrotic scars, particularly in the alveolar region, leading to reduced gas exchange and chronic respiratory failure.39 Based on the above-mentioned damage Pneumocystis can cause in the lungs, the pathogenesis of P. carinii infection is, thus, of great significance.
Fibronectin (Fn) is an extracellular macromolecular membrane protein found on the surface of animal cells. It is the major non-collagenous glycoprotein in the extracellular matrix (ECM) and basement membrane. Fn1, a member of the Fn family, is involved in a variety of cell biological processes, such as embryogenesis, wound healing, blood clotting, host defense, metastasis, cell adhesion and migration during cell proliferation.20 Besides, Fn1 is involved in fibrosis and other diseases.28 Studies have shown that the more severe the bladder fibrosis, the higher the level of Fn1 expression.13 Su et al.30 developed a competing endogenous RNA (ceRNA) associated with renal fibrosis, the analysis of which revealed that extracellular Fn1 deposition may be regulated by the transcription factor Creb5. Chen et al.3 found that when lung fibrosis in mice was reduced, Fn1 expression decreased. Although the relationship between Fn1 and tissue fibrosis has been reported previously, the mechanism of P. carinii infection-induced pulmonary fibrosis remains to be explored. Therefore, it is essential to investigate the molecular mechanisms involved in P. carinii infection.
In this study, we performed a series of bioinformatics analyses on mice infected with P. carinii, screened the Fn1 with the highest degree of connectivity through a protein–protein interaction (PPI) network, and investigated the regulatory role of the Fn1 in pulmonary fibrosis and inflammation. The results were conducive to the interpretation of pulmonary fibrosis development.
Materials and methodsDataset accessmRNA chip data of GSE20149 (platform GPL85) dataset were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/), which contained four alveolar macrophages samples of normal mice (Mus musculus) and another four from mice infected with P. carinii.
Screening of gene differentially expressed after P. carinii infectionWith the normal samples as the control, the GSE20149 microarray data were analyzed by the R package “limma”.21 The differentially expressed mRNAs (DEmRNAs) after the mice were infected with P. carinii (|logFC|>1.5, padj<0.05) were screened.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysesIn order to explore the biological functions of these DEmRNAs, the “clusterProfiler” package was used for GO and KEGG enrichment analyses (q value <0.05, p value <0.05).42
PPI construction and analysisAfter inputing DEmRNAs to STRING website (https://string-db.org/cgi/network?taskId=bCtYmfq6AVjK&sessionId=bn6bKqAwpP27) for PPI network analysis, confidence score was set greater than or equal to 0.7, and the connectivity of genes in the network was calculated to determine the target mRNA. Wilcoxon was used to examine the difference of target mRNA expression between normal samples and samples infected with P. carinii (p value <0.05).
Development of a Fn1 knockout mouse modelWe developed a mouse model of Fn1 deletion-alveolar epithelial cells following experimental methods described by Gui et al.8 and Ouyang et al.18 In short, Fn1flox/floxC57BL/6J mice were hybridized with SPCCre/+C57BL/6J mice to obtain mice with Fn1 deletion-alveolar epithelial cell. Mice with Fn1 deletion were named SPC-Cre:Fn1fl/fl, and the control was named Fn1fl/fl. Pathogen-free C57BL/6 mice were purchased from Laboratory Animal Center of Chinese Academy of Sciences in Shanghai (Shanghai, China).
Development of mouse models infected with P. cariniiThe mouse model of PCP was developed according to the method described by Cushion et al.4 The experiment was conducted in four groups: Vehicle+Fn1fl/fl group, Vehicle+SPC-Cre:Fn1fl/fl group, PCP+Fn1fl/fl group and PCP+SPC-Cre:Fn1fl/fl group. The PCP model was developed by injecting subcutaneously in the groin the immunosuppressant dexamethasone (0.02mg/g) (D4902-25MG, MERCK, USA) every two days for 45 days. At the same time, cold-boiled water with 1mg/mL tetracycline hydrochloride was prepared as drinking water for each group to prevent any bacterial infection. There was no limitation on the amount of drinking water. The weight of each mouse was recorded every day during the treatment, and their general condition was also observed. After 45 days, all mice were killed, and their lungs were weighed. The lung tissue was fixed in 4% paraformaldehyde for 24 h, and then frozen for subsequent experiments. All procedures were reviewed and approved by the Animal Ethics Committee.
Histopathological examinationAfter fixing lung tissues with 4% paraformaldehyde and embedding them in paraffin, we cut the paraffin block in 5μm-thickness sections using a microtome. Sections were stained with hematoxylin and eosin (H&E), or Masson trichromatic staining (Sinopharm Chemical Reagents Beijing Co., Ltd., China). The inflammation and collagen deposition were observed under the optical microscope.
Calculation and assessment of lung coefficient and hydroxyproline levelLung coefficient refers to the weight of the lungs divided by the weight of the body. Hydroxyproline is the indicator of collagen level in the lung tissue of mice. These two indicators can also be used to estimate pulmonary fibrosis degree in mice.14 After pretreating lung tissue, the hydroxyproline level in the lung was assessed by spectrophotometry using the hydroxyproline detection kit (Nanjing Jiancheng Bioengineering Institute, China) according to the manual. Results were presented as hydroxyproline per milligram of wet lung (μg/mg).
Immunofluorescence and immunohistochemistry (IHC)Paraffin-embedded lung tissue sections were first stained with anti-Fn1 antibody (ab2413; Abcam, UK) and then stained with goat anti-rabbit IgG (ab150077; Abcam, UK) with Alexa Fluor™ 488. After that, DAPI (Invitrogen, USA) was added to counterstain the nucleus. Finally, the sections were observed under a fluorescent microscope.40
IHC staining was performed using an anti-Fn1 antibody (26836S; Cell Signaling Technology, USA), anti-F4/80 antibody (ab300421, Abcam, UK), anti-CD45 antibody (ab10558, Abcam, UK) and anti-α-SMA antibody (ab5649, Abcam, UK), according to the methods mentioned in a previous study.32
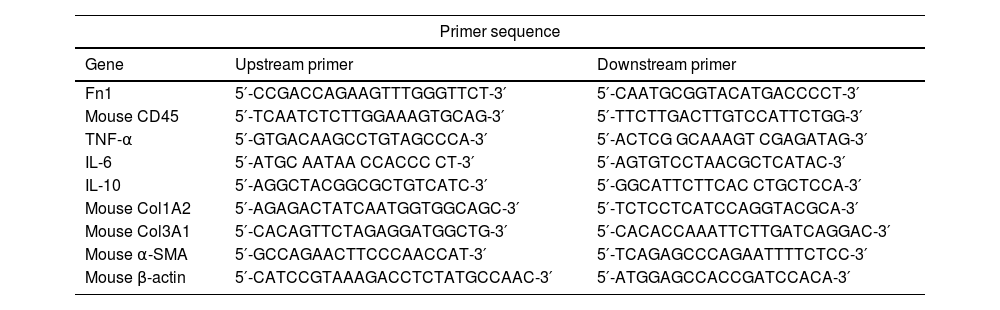
Real-time quantitative polymerase chain reaction (qRT-PCR)The total RNA in the lung tissue of each mice was extracted by the Trizol method, and cDNA was synthesized using the reverse transcription kit (Applied Biosystems, USA). Leverage PowerUp™ SYBR® Green Master Mix (Invitrogen, USA) was employed to detect the relative expression of target genes. The specific primer sequences are shown in Table 1.
Primer sequence.
| Primer sequence | ||
|---|---|---|
| Gene | Upstream primer | Downstream primer |
| Fn1 | 5′-CCGACCAGAAGTTTGGGTTCT-3′ | 5′-CAATGCGGTACATGACCCCT-3′ |
| Mouse CD45 | 5′-TCAATCTCTTGGAAAGTGCAG-3′ | 5′-TTCTTGACTTGTCCATTCTGG-3′ |
| TNF-α | 5′-GTGACAAGCCTGTAGCCCA-3′ | 5′-ACTCG GCAAAGT CGAGATAG-3′ |
| IL-6 | 5′-ATGC AATAA CCACCC CT-3′ | 5′-AGTGTCCTAACGCTCATAC-3′ |
| IL-10 | 5′-AGGCTACGGCGCTGTCATC-3′ | 5′-GGCATTCTTCAC CTGCTCCA-3′ |
| Mouse Col1A2 | 5′-AGAGACTATCAATGGTGGCAGC-3′ | 5′-TCTCCTCATCCAGGTACGCA-3′ |
| Mouse Col3A1 | 5′-CACAGTTCTAGAGGATGGCTG-3′ | 5′-CACACCAAATTCTTGATCAGGAC-3′ |
| Mouse α-SMA | 5′-GCCAGAACTTCCCAACCAT-3′ | 5′-TCAGAGCCCAGAATTTTCTCC-3′ |
| Mouse β-actin | 5′-CATCCGTAAAGACCTCTATGCCAAC-3′ | 5′-ATGGAGCCACCGATCCACA-3′ |
The cells were treated with RIPA lysis buffer (Thermo Fisher Scientific, USA), and the protein concentration was determined with BCA assay kit (Thermo Fisher Scientific, USA). Once separated by SDS-PAGE, proteins were transferred to PVDF membrane, sealed with 5% skimmed milk at room temperature for 1h, and incubated with the first antibody at 4°C overnight. After three times of 1× TBST wash, lasting 10min each, proteins were incubated with the second antibody (goat anti-rabbit horseradish peroxidase – HRP, 1:2000, ab6721; Abcam, UK) for 1h at room temperature. Enhanced chemiluminescence (ECL) kit (Thermo Fisher Scientific, USA) was used to observe and analyze bands. Primary antibodies used in this study included anti-Fn1 (26836S; Cell Signaling Technology, USA), anti-NLRP3 antibody (ab263899, Abcam, UK), anti-α-SMA (19245S; Cell Signaling Technology, USA), anti-GAPDH (ab8245, Abcam, UK), and anti-β-actin (ab115777; Abcam, UK).
Enzyme-linked immunosorbent assay (ELISA)Mouse blood samples were collected, and supernatant was obtained by centrifuging the blood at 1000g for 20min at 4°C after natural coagulation at room temperature. The concentration of five cytokines in serum samples was measured by ELISA using commercially available kits according to the manufacturer's instructions. The concentration of cytokine or chemokine was calculated based on standard curves provided with the kits, and results were expressed in pg/ml. Mouse tumor necrosis factor alpha (TNF-α) ELISA kit (ab208348), mouse interleukin-6 (IL-6) ELISA kit (ab222503), mouse interleukin-10 (IL-10) ELISA kit (ab255729), mouse interleukin-1 beta (IL-1β) ELISA kit (ab197742) were purchased from Abcam (UK) and mouse interleukin-8 (IL-8) ELISA kit (ml058632) was purchased from MLBio (China).
Statistical analysisAll statistical analyses were performed on GraphPad Prism 8 software (GraphPad Software, Inc., La Jolla, USA). All experiments were repeated at least three times. The results were expressed as mean±standard deviation. The data between groups were compared by t-test or one-way analysis of variance. Differences were statistically significant when p<0.05.
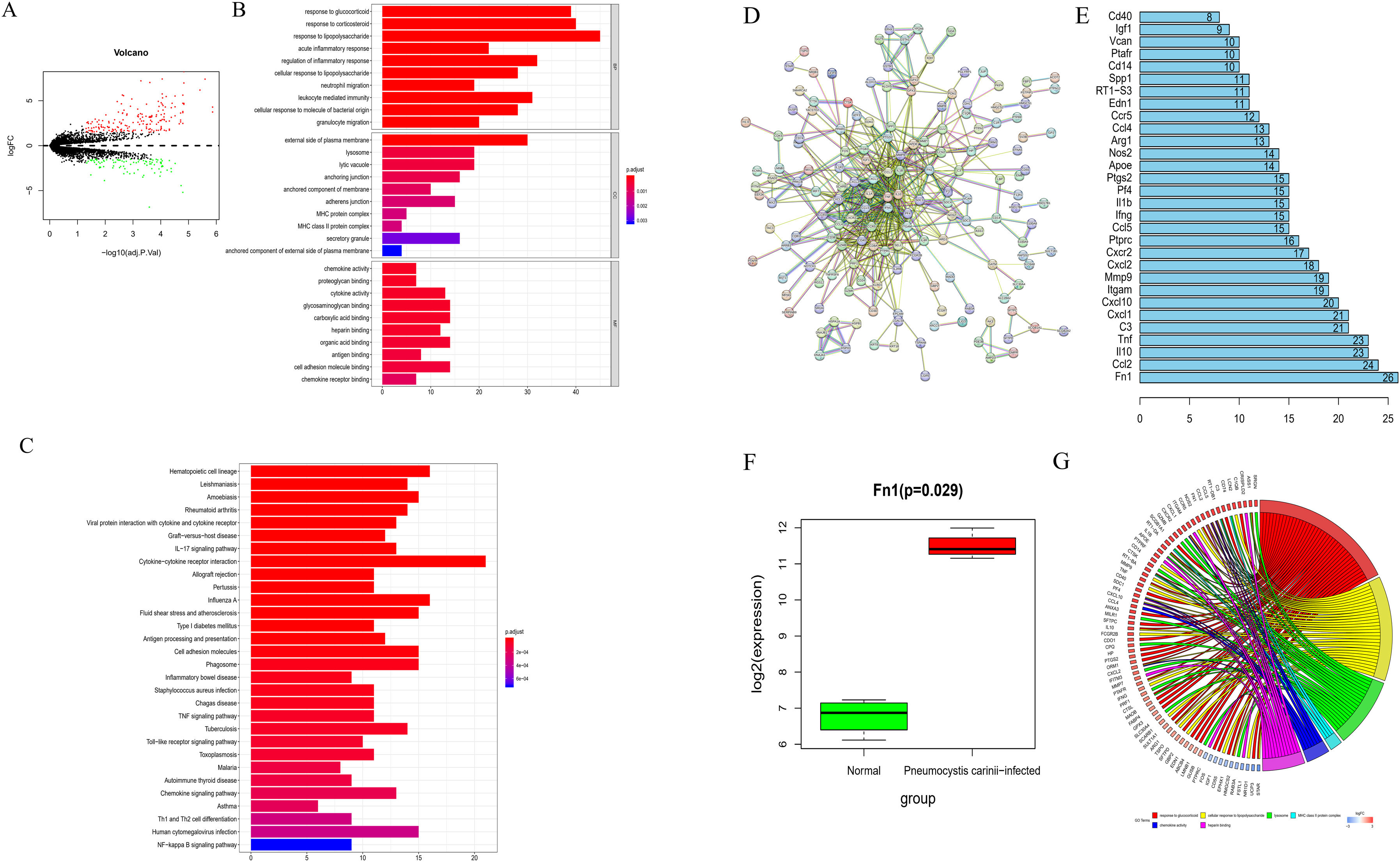
ResultsDEmRNAs screening and functional enrichment analysisThrough differential analysis of alveolar macrophages samples from normal mice and mice infected with P. carinii in the GSE20149 dataset, 273 DEmRNAs were obtained, including 179 up-regulated mRNAs and 94 down-regulated mRNAs (Fig. 1A).
Biological function analysis of mice infected with P. carinii. (A) Volcano map of DEmRNAs between normal group and P. carinii-infected group from GSE20149 dataset; (B) GO enrichment analysis of DEmRNAs; (C) KEGG enrichment analysis of DEmRNAs; (D) PPI network based on DEmRNAs; (E) the degree value diagram, the X-axis represents the degree value, and the Y-axis represents the gene name; (F) the expression of Fn1 in the GSE20149 dataset. The green box diagram represents the normal samples, and the red box diagram represents the samples infected with P. carinii; G. GO enrichment analysis.
In order to analyze the biological function regulated by these DEmRNAs, GO and KEGG enrichment analyses were carried out. GO analysis results showed that these DEmRNAs were significantly enriched in the acute inflammatory response, regulation of inflammatory response, leukocite mediated immunity and other functions related to immunity and inflammation. The results of KEGG analysis indicated that these genes were significantly enriched in the signal pathways related to inflammation, such as TNF signaling pathway and Toll-like receptor signaling pathway (Fig. 1B and C).
The PPI network was developed using the STRING website (Fig. 1D). After analyzing statistically the topological properties of the network (by degree), we ranked the top 30 hub genes and found that Fn1 had the highest degree of connectivity in the entire network (Fig. 1E). Compared with the samples from non-infected mice, the ones from mice infected with P. carinii showed upregulation of Fn1 (Fig. 1F). Subsequently, we used GO enrichment analysis to investigate the signal pathway affected by Fn1. According to the results, Fn1 was involved in biological processes such as response to glucocorticoid and cellular response to lipopolysaccharide (Fig. 1G). Based on the above results, we speculate that the infection by P. carinii may aggravate the inflammatory reaction and promote the progression of pulmonary fibrosis by increasing the expression of Fn1.
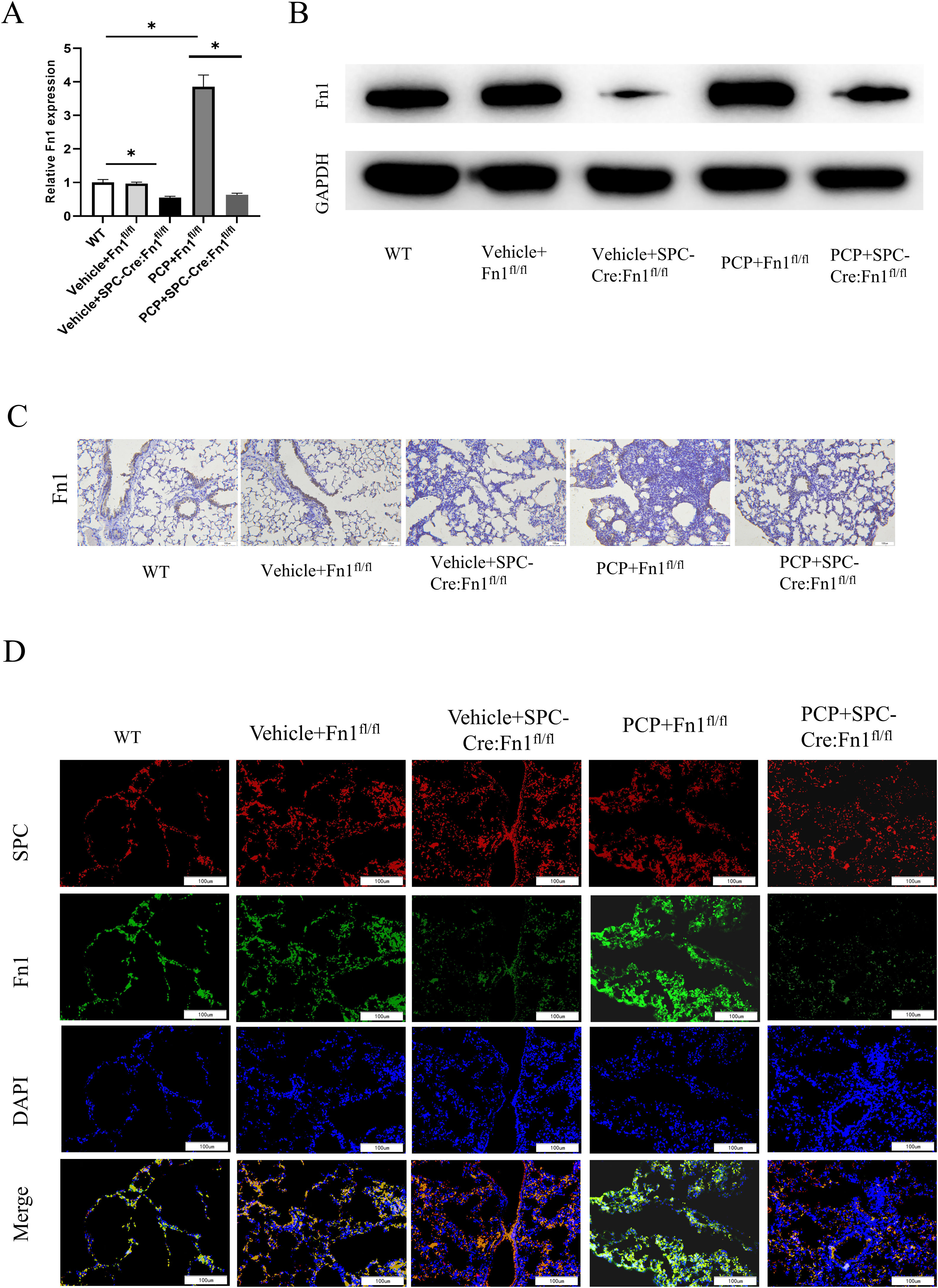
Validation of Fn1 expression in mice lung tissuesWe observed that mice in the wild type (WT), Vehicle+Fn1fl/fl group, and Vehicle+SPC-Cre:Fn1fl/fl group exhibited normal general conditions, regular diet and activity, shiny coat, significant weight gain, and no apparent lung lesions upon dissection. However, mice in the PCP+Fn1fl/fl group showed reduced activity and food intake from the fourth week, dull and lackluster coat, lethargy, clustering in corners, gradual weight loss, hazy and darkened eyeballs with inflamed secretions, and rapid breathing. Dissection of the lungs revealed scattered nodular lesions of varying sizes, some merging into patches, with lung atrophy, hardened texture, and a grayish-white appearance. Mice in the PCP+SPC-Cre:Fn1fl/fl group exhibited milder symptoms compared to those in the PCP+Fn1fl/fl group. To explore the regulatory effect of PCP on Fn1, we used qRT-PCR, WB and IHC to detect the expression of Fn1. The results found in the PCP+Fn1fl/fl group was not significantly different from that in WT group, and the expression level of Fn1 in the Vehicle+SPC-Cre:Fn1fl/fl group was the lowest (Fig. 2A–C), indicating the successful development of the Fn1 knockout mouse model. In addition, compared with Vehicle+Fn1fl/fl group, PCP+Fn1fl/fl group had a significantly higher Fn1 expression. Subsequently, the expression of Fn1 was verified by immunofluorescence assay, the result of which coincided with the above results (Fig. 2D). These observations led to the conclusion that dexamethasone treatment increased the expression of Fn1 in Fn1fl/fl mice, but had no effect on Fn1 expression in SPC-Cre:Fn1fl/fl mice.
Validation of Fn1 expression in mice lung tissues. (A) The expression of Fn1 in each group was detected by qRT-PCR; (B) the expression of Fn1 protein in each group was detected by WB; (C) the expression of Fn1 protein in each group was detected by IHC; (D) immunofluorescence staining results of Fn1 in each group. *p<0.05.
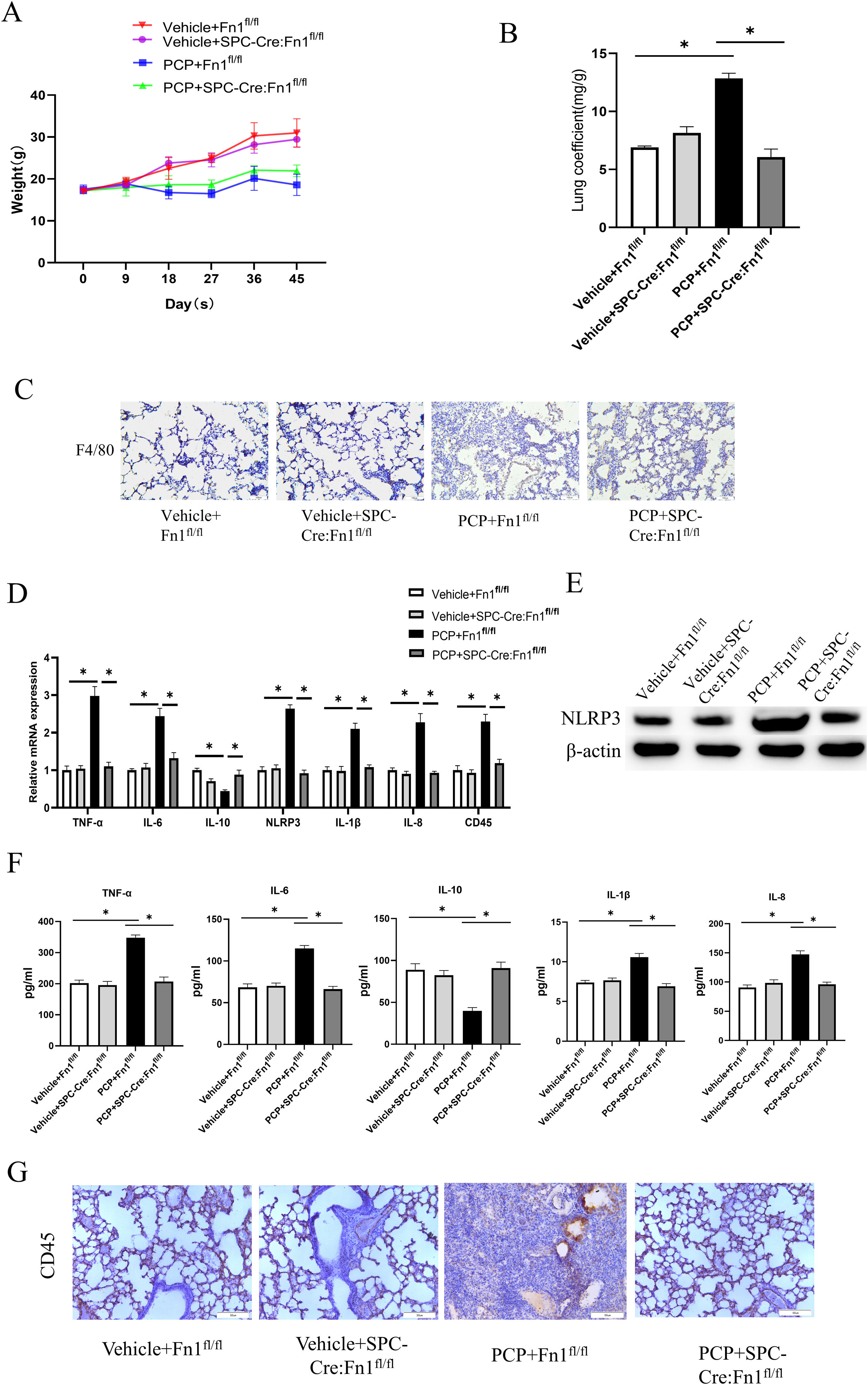
To verify the relationship between Fn1 and pulmonary fibrosis, we assessed lung injury in mice. Compared with the Vehicle+Fn1fl/fl group, PCP+Fn1fl/fl group showed weight loss within 45 days (Fig. 3A). The mice weight in PCP+SPC-Cre:Fn1fl/fl group depleted of Fn1 was higher than that of the PCP+Fn1fl/fl group. After 45 days of infection, the highest lung coefficient was found in the PCP+Fn1fl/fl group, which indicated that the mice had serious lung injury (Fig. 3B).
Measurement of lung injury and inflammation magnitude in mice. (A) Weight changes; (B) lung coefficient; (C) macrophage infiltration was detected by IHC; (D) TNF-α, IL-6, IL-10, IL-1b, IL-8, NLRP3 and CD45 expression levels were detected by qRT-PCR; (E) NLRP3 expression level was detected by WB; (F) TNF-α, IL-6, IL-10, IL-1b and IL-8 expression levels were detected by ELISA; (G) Expression of CD45 in lung tissue of mice in each group was detected by IHC. *p<0.05.
Subsequently, to determine the effect of Fn1 on the degree of lung inflammation in mice, we examined macrophage infiltration using IHC. That infiltration is attributed to the large amounts of inflammatory cytokines produced by macrophages that trigger a cascade response of inflammatory mediators, leading to massive tissue destruction.9 The results showed that macrophage infiltration in the PCP+Fn1fl/fl group was significantly increased after stimulation with dexamethasone, while it was alleviated in the PCP+SPC-Cre:Fn1fl/fl group (Fig. 3C). Then, we further examined the expression of inflammatory factors. According to the qRT-PCR results, after dexamethasone treatment in the PCP+Fn1fl/fl group, the expression of the pro-inflammatory factors TNF-α, IL-6, IL-1β and IL-8 was up-regulated, the expression of NLRP3 inflammasome was also up-regulated, and the expression level of the anti-inflammatory factor IL-10 decreased significantly. These changes were alleviated in the PCP+SPC-Cre:Fn1fl/fl group (Fig. 3D). The expression of NLRP3 protein was detected using WB (Fig. 3E), and the expression level of cytokines in serum was tested using ELISA (Fig. 3F). The results showed that compared with the Vehicle+Fn1fl/fl group, NLRP3 protein expression and serum levels of TNF-α, IL-6, IL-1β and IL-8 were also significantly higher in the PCP+Fn1fl/fl group, while IL-10 levels were significantly lower, whereas these indices did not have significant changes in the PCP+SPC-Cre:Fn1fl/fl group, in which the protein expressions were similar to the trend seen in Fig. 3D. Next, the expression of CD45 was detected. qRT-PCR and IHC results showed a significant increase in CD45 expression in the PCP+Fn1fl/fl group, whereas there was no significant change in the PCP+SPC-Cre:Fn1fl/fl group (Fig. 3D and G). In addition, no significant difference was observed between the Vehicle+Fn1fl/fl group and the Vehicle+SPC-Cre:Fn1fl/fl group in the above-mentioned validation experiments. The above results suggest that the absence of Fn1 led to negligible changes in lung conditions if no dexamethasone was administered. Nevertheless, if combined with dexamethasone treatment, upregulation of Fn1 resulted in severe lung injury and inflammation. After silencing Fn1, the lung injury and inflammation were relieved.
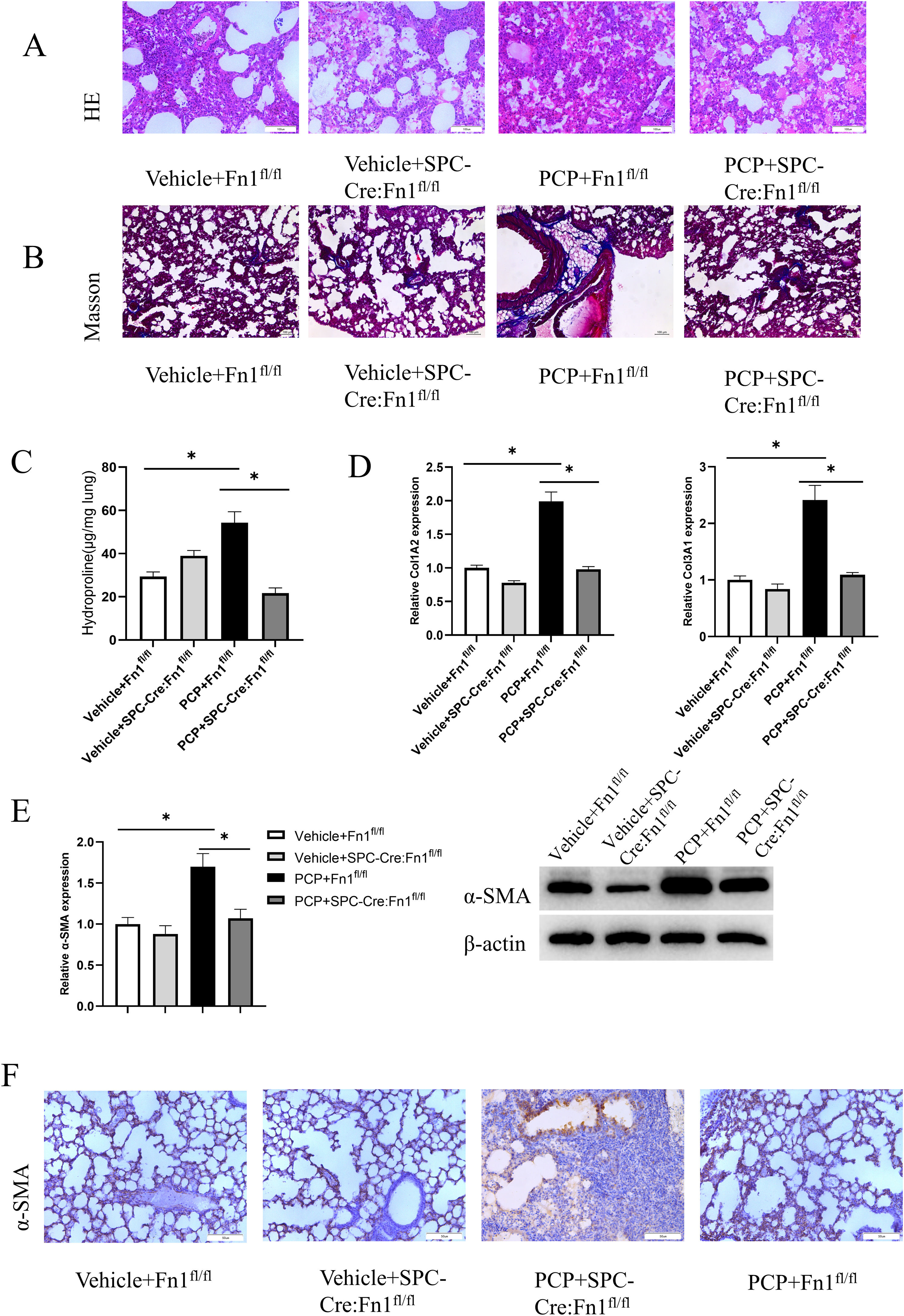
Pulmonary fibrosis assessment in miceTo detect the lung fibrosis degree in each group, we carried out H&E staining (Fig. 4A) and Masson staining (Fig. 4B) after 45 days of treatment. The strongest staining happened in PCP+Fn1fl/fl group. PCP+SPC-Cre:Fn1fl/fl group displayed a lighter staining. Similarly, the highest hydroxyproline level was found in PCP+Fn1fl/fl group, indicating a more severe pulmonary fibrosis. However, PCP+SPC-Cre:Fn1fl/fl group presented the lowest level of hydroxyproline in lung tissue, which suggested that the overexpressed Fn1 could promote the pulmonary fibrosis in mice (Fig. 4C).
Pulmonary fibrosis degree assessment in mice. (A) H&E staining of lung tissue in each group; (B) Masson staining of lung tissue in each group; (C) hydroxyproline level; (D) content determination of collagen Col1A2 and Col3A1; (E) RNA level and protein level of a-SMA; (F) a-SMA expression detected by IHC experiment. *p<0.05.
Apart from that, we found that the mice in PCP+Fn1fl/fl group displayed the highest expression of Col1A2 and Col3A1, indicators of collagen level, suggesting the most severe degree of pulmonary fibrosis (Fig. 4D). When assessing α-SMA in myofibroblasts, it was found that compared with the Vehicle+Fn1fl/fl group, the PCP+Fnfl/fl group had increased levels of RNA and protein α-SMA. The mice in PCP+SPC-Cre:Fn1fl/fl group had decreased levels of RNA and protein α-SMA when compared with the PCP+Fn1fl/fl group (Fig. 4E). The same results were also found in the IHC assay (Fig. 4F). The above results show that Fn1 knockout significantly inhibited the formation of pulmonary fibrosis in mice.
DiscussionThough medical advances have improved survival, neither the treatment, nor PCP prognosis, have experienced significant progress. In recent years, the use of immunosuppressants and the spread of chemotherapy in cancer have placed premature and malnourished infants, as well as cancer and HIV patients, at the highest risk category of PCP.22 Pneumonia fibrosis has been described in HIV patients after receiving PCP treatment.31 Pulmonary fibrosis is characterized by the replacement of normal alveolar structures by collagen-rich matrix, which is a common pathological response to lung injury.36 Pharmacological interventions do not reverse the progression of pulmonary fibrosis, which leads to severe lung damage and even death.17 To date, limited therapeutic strategies for pulmonary fibrosis remains a daunting challenge for both clinicians and researchers.
Fibrotic tissue is characterized by the excessive accumulation of extracellular matrix (ECM) proteins. As an important ECM protein in the development of fibrosis, Fn1 mediates in the interaction between various ECM components and cells,43 as well as in the assembly of other ECM components.26 For example, the Fn1 matrix is essential in the formation of collagen networks.35 The cells responsible for collagen production are thought to be fibroblasts and lung fibroblasts.5 The number of lung fibroblasts and fibroblasts were linked with the progression of collagen deposition and pulmonary fibrosis in patients. As the major cause of pulmonary fibrosis, fibroblasts can take a huge toll on lung function when excessively increased.24 Upregulated during lung fibroblasts, Fn1 was found to play an important role in TGF-β-induced myofibroblast differentiation.34 The finding was consistent with our observation that Fn1 expression was significantly increased in mice following dexamethasone-induced pulmonary fibrosis, and was significantly reduced following the knockdown of Fn1.
Collagen is a major component of the ECM. In most cases, the secretion and degradation of ECM by fibroblasts are in a dynamic balance, which would be disrupted if pulmonary fibrosis occurs.15 In addition, increased collagen may promote inflammation.19 Scholars have demonstrated through a variety of studies that promoting inflammation can exacerbate the formation of pulmonary fibrosis. Besides, the inflammatory response and pulmonary fibrosis are relieved when following pharmacological treatment.6,12 IL-10, an anti-inflammatory mediator, has become one of the key research targets for potential anti-fibrotic therapy.27 It was reported in a study by Fei et al.11 that bleomycin-induced lung fibrosis in mice could be reduced by modulating NF-κB/TNF-α signaling in macrophages. Apart from that, Xie et al.23 developed a mouse model of dexamethasone-induced lung injury and found that increased IL-6 serum levels were associated with pulmonary fibrosis. A recent study demonstrated that the activation of NLRP3 inflammasome can promote pulmonary fibrosis in rats.38 Multiple studies have reported the involvement of the pro-inflammatory cytokine IL-1β in the pathogenesis of pulmonary fibrosis.2,25 Bhusal et al.1 found that macrophages release a large amount of IL-8 to recruit other leukocytes to the site of infection, thereby triggering an inflammatory response. Additionally, IL-8 has been shown to promote idiopathic pulmonary fibrosis.41 Similar to previous reports, this study found that the expression levels of the pro-inflammatory factors TNF-α, IL-6, IL-1β and IL-8 were significantly higher, the NLRP3 inflammasome was activated, and the expression level of anti-inflammatory factor IL-10 was significantly lower in the lung tissue of dexamethasone-treated mice than in normal mice. In Fn1-silenced mice, the level of inflammation was significantly reduced, indicating that Fn1 could promote lung fibrosis and inflammation.
In this study, through screening of DEmRNAs in PCP, functional enrichment analysis and PPI network construction, we found significantly high expression of Fn1 in lung samples of P. carinii-infected mice. A series of in vivo and molecular experiments then confirmed that P. carinii infection drove the upregulation of Fn1, which in turn caused pulmonary fibrosis with an inflammatory response. Nevertheless, there were still some limitations in our study, including the unknown specific regulatory mechanism of Fn1 in pulmonary fibrosis and the absent experimental validation of its downstream regulatory genes. In conclusion, this study provides some insights into the mechanisms of lung fibrosis development.
Ethical approvalThe study was conducted after being approved by the Animal Ethics Committee of Affiliated Yueqing Hospital, Wenzhou Medical University (Number: YQYY202300005).
FundingNot applicable.
Authors’ contributionsWenwen Yu, Hua Ye and Yunlei Li conceived and designed the study.
Xiaoqiong Bao, Yangyang Ni, Xiangxiang Chen and Yangjie Sun performed the experiments.
Ali Chen, Weilong Zhou and Jifa Li wrote the paper.
All authors read and approved the manuscript.
Availability of data and materialsThe data and materials in the current study are available from the corresponding author on reasonable request.
Competing interestThe authors declare no conflicts of interest.