Clinical HER2 positive (cHER2+) breast cancer (BC) is an heterogenous disease, which represents a major challenge in the design of personalized therapeutic strategies. In recent years, a great development against HER2 targeted therapies took place, but not all patients benefit from the same regimens. In this context, it is essential to develop genomic analysis, with clinicopathological and molecular biomarkers, as HER2DX, in order to improve response, safety, and patient's quality of life. This review aims to comprehensively summarize the current evidence regarding the development, validation, and application of HER2DX score in clinical practice. Genomic development and clinical application in the identification of different cHER2+ BC populations are discussed, along as future changing strategies for escalation and de-escalation treatments.
El cáncer de mama (CM) HER2 positivo (cHER2+) es una enfermedad muy heterogénea, lo que representa un gran reto en el diseño de estrategias terapéuticas personalizadas. En los últimos años, se ha producido un gran desarrollo en las terapias dirigidas anti-HER2, pero no todas las pacientes se benefician de los mismos esquemas de tratamiento. Por ello, es fundamental desarrollar test genómicos, con biomarcadores clinico-patológicos y moleculares, como HER2DX, para mejorar la respuesta, la seguridad y la calidad de vida de las pacientes. El objetivo de esta revisión es resumir de forma exhaustiva la evidencia actual sobre el desarrollo, la validación y la aplicación de HER2DX en la práctica clínica. Además, se discute el desarrollo genómico y la aplicación clínica en la identificación de diferentes poblaciones de CM cHER2+, así como las futuras estrategias de cambio para los tratamientos de escalada y desescalada.
Clinical HER2 positive (cHER2+) disease has a prevalence around 15% of all breast cancer (BC) and is associated with poor prognosis.1 However, in the last decades, thanks to the development of innovative therapies such as anti-HER2 Antibody Drug Conjugated (ADCs), the prognosis of HER2-positive mBC significantly improved with median overall survival (OS) of 57 months.2 In 80% of cases, it is characterized by the amplification of ERBB2 gene, located on chromosome 17q12, that encodes a tyrosin kinase receptor (TKR) responsible of the activation of several signaling pathways.3–6 HER2 status is determined by combining immunohistochemistry (IHC) techniques and in situ hybridization (ISH) on formalin-fixed paraffin-embedded (FFPE) biopsies derived from primary BC or recurrent/metastatic tumors.6 HER2+ is defined by an IHC score of 3+ or 2+ with performance of ISH and subsequent results: ERBB2/CEP17 ratio ≥ 2.0 and ≥4.0 ERBB2signal/cell; ERBB2/CEP17 ratio < 2.0 and ≥6.0 ERBB2signals/cell; or if the average ERBB2 copy number is ≥6.0 signals/cell.7–9
cHER2+ BC is an heterogeneous disease, tumors with similar morphology and stage may exhibit variations in clinical behavior and response to treatment, depending on their molecular characteristics. The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets demonstrated that all the intrinsic molecular subtypes can be identified within HER2+ disease, although HER2 enriched (HER2-E) is the most frequent (∼47.0%), followed by Luminal B (∼18–28.2%), Luminal A (11–23%), and Basal-like (7–14%).3,4,10 For a given molecular subtype, there are no biological differences between cHER2+ and cHER2-negative disease, except for the expression of genes close to ERBB2 amplicon.3,10
As a clinical variable, HER2 is an independent prognostic factor, but this relevance disappears in molecular classification, where is not related to major changes in signaling pathways or survival.3,10
In cHER2+/HER2-E disease, ERBB2 amplification appears as a main driver in 80% of cases, showing the highest activation of the pathway and the greatest impact to double antiHER2 therapy. HER2-E tumors have increased expression of FGFR4, EGFR, and ERBB2 amplicon genes (GRB7 y PGAP3), while tumors characterized by hormone receptor positive (HR+) status are marked by PIK3CA mutations (40%) and increased expression of GATA3, BCL2, and ESR1 genes.3–5,8,11
In recent years, targeted therapies against HER2 early breast cancer (eBC) disease changed the prognosis of patients, but not all of them benefit from the same therapy regimens. Currently, there are no biomarkers that allow treatments to be personalized, so decisions to de-escalate and escalate treatments are based on clinicopathological factors. Given the urgent need of a prognostic tool for these tumors, HER2DX test has been developed.
Material and methodsTo carry out this work, we made a bibliographic search of high impact scientific articles published in international journals, consulting PubMed database. This bibliography includes systematic reviews, clinical trials, retrospective studies, and conference communications, published in the last 12 years (2012–2024).
ResultsHER2DX 13-gene testHER2DX 13-gene test was the first developed model using retrospective, clinical, pathological, and genomic data from 435 patients in the Short-HER trial,12,13 a phase III study comparing 9 weeks vs 1 year of adjuvant trastuzumab combined with chemotherapy.14
After development, HER2DX 13-gene test was evaluated retrospectively in a cohort of patients from 2 trials and 2 European hospitals. The CHER-LOB trial included 74 patients treated with preoperative chemotherapy plus trastuzumab, trastuzumab plus lapatinib or lapatinib alone.15 From PAMELA study, they evaluated 88 patients treated with neoadjuvant lapatinib and trastuzumab.16 The 2 cohorts from Hospital Clinic and Padova University evaluated 68 and 37 patients, respectively, treated with neoadjuvant chemotherapy and trastuzumab.12,13
HER2DX final model included 17 variables: nodal stage (N0 vs N+), tumor size (T1 vs T ≥ 2), MMP11, PAM50 subtype (HER2-E and basal-like vs rest), and 13 individual genes.12 Of these 13 genes, 7 were associated with poor distant metastasis-free survival (DMFS) (CDC6, EXO1, RRM2, TMEM45B, FGFR4, and CDH3) and other 6 with a good outcome (BAG1, KRT5, KRT4, MLPH, MYC, and PHGDH).12 Tested as a continuous variable, HER2DX low-risk showed longer DMFS compared to medium, medium-high, and high-risk groups.12 HER2DX risk score was not associated with pathological complete response (pCR) (OR 1.02, 95% CI 0.6–1.6, P = .93) and patients without a pCR in the HER2DX low-risk group had longer disease-free survival (DFS) compared to the other subgroups.12
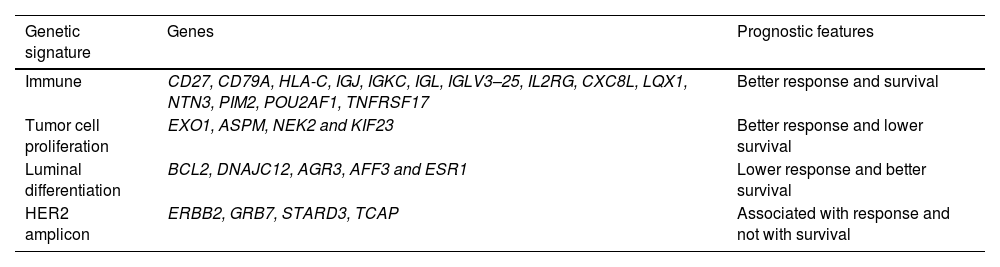
HER2DX 27-gene testThe first version of HER2DX did not provide information about pCR after neoadjuvant treatment and a new version with 2 independent scores, long-term prognostic risk score (HER2DX risk score) and pCR likelihood score (HER2DX pCR score) was developed.13 This new version includes clinical features and 27 genes expression grouped into 4 genomic signatures, that recently demonstrated precision, reproducibility, and low variability across laboratories and platforms.13,17 Clinical features were tumor stage (T1 vs T2 vs T3–4) and nodal status (N0 vs N1 vs N2–3).13 The 4 genomic signatures were immune (CD27, CD79A, HLA-C, IGJ, IGKC, IGL, IGLV3–25, IL2RG, CXC8L, LQX1, NTN3, PIM2, POU2AF1, and TNFRSF17), tumor cell proliferation (EXO1, ASPM, NEK2, and KIF23), luminal differentiation (BCL2, DNAJC12, AGR3, AFF3, and ESR1) and HER2 amplicon (ERBB2, GRB7, STARD3, and TCAP).13
HER2DX risk scoreHER2DX risk score was developed in 434 patients from Short-HER trial, where it demonstrated a significant association with disease recurrence-free survival (DRFS) (P < .0001).13 Two risk groups, range from 0 to 100, were defined as low risk (0–50) and high risk (50–100).13
Subsequently, validation was carried out in 147 patients from Hospital Clinic, 84 from PAMELA trial, and 37 from Padova University, where HER2DX risk score showed a significantly association with DFS.13 HER2DX low-risk had significantly longer DFS at 5 years than high risk (97.4% vs 84.7%, respectively; HR 0.21, 95% CI 0.1–0.6, P = .005).13 In public dataset (TCGA, METABRIC, SCAN-B, and CALGB-4060I) a statistically significant association with OS was observed.13
Dieci et al. hypothesized that integration of molecular profiles, intrinsic subtypes, and HER2DX score, with clinicopathological characteristics, could help to recognize HER2+ eBC at increased risk of relapse in specific sites.18 They analyzed 677 patients and they found that HER2DX risk score was associated with increased risk of all-site metastasis relapse, with the exception of the brain: 10 year cumulative incidence of 19.7% vs 5.3% for high vs low risk score (P < .001).18 The incidence of brain metastasis was statistically significantly higher for HER2-E subtype in patients with HER2DX high risk score.18
Recently, HER2DX risk score demonstrated association with OS after adjustment by clinical variables and treatment regimens (7-year OS rate of 94.5% in HER2DX low vs 78.6% in HER2DX high; HR 3.87, 95% CI 2.26–6.65, P < .001).19
HER2DX pCR scoreHER2DX pCR score was developed in 116 patients from Hospital Clinic and further validation was carried out in 2 cohorts from PAMELA trial (91 patients) and Hospital Clinic/Padova (67 patients). Two variables were associated with pCR (immune and proliferation) and other 2 with non-pCR (luminal tumor and nodal staging). A significant relation between HER2DX pCR score and pCR was demonstrated (P < .0001) and 3 subgroups, range from 0 to 100, (low = 0–33.3; medium = 33.3–66.7; high = 66.7–100) were established.13 Public dataset (CALGB-4060I and ISPY-2) also observed an association between HER2DX pCR score and pCR.13
Villacampa et al. tested the ability of HER2DX pCR score to predict pCR across different treatment subgroups. In HER2DX pCR-high, they found a significant increase in pCR rate due to HER2 blockade (OR = 2.36, 95% CI 1.09–5.42, P = .03) and among patients with HER2DX pCR-medium score due to multi-agent chemotherapy (OR = 3.11, 95% CI 1.54–6.49, P = .002).20
In daily clinical practice, HER2DX pCR test also revealed an association with pCR (81% in pCR-medium/high score and 32% in pCR-low score; OR = 9.3, P = .001).21
HER2DX ERBB2 mRNA scoreERBB2 expression within HER2+ disease can help to identify patients with higher response to anti-HER2 therapy.13,22 HER2-E subtype, with ERBB2 mRNA high expression, showed a pCR of 45% to neoadjuvant lapatinib plus trastuzumab, which reflects the importance of ERBB2 mRNA levels to predict anti-HER2 sensitivity and justify the development of a test for validation as a biomarker.22
During development of HER2DX test, Prat et al. revealed that HER2DX ERBB2 mRNA assay predicts cHER2 status with 84% sensitivity and 100% specifity.12,21 No cHER2-negative cases were identified as ERBB2+ and 16.4% of cHER2+ were identified as ERBB2-negative/low.13
Bertucci et al. analyzed ERBB2 expression in 127 cHER2+ patients treated in I-SPY2 trial with neoadjuvant chemotherapy plus anti-HER2 blockade vs T-DM1 and pertuzumab followed by doxorubicin and cyclophosphamide.24 They found that ERBB2 mRNA expression was associated with pCR in both cohorts, but OR was higher in T-DM1 group (OR 7.09, 95% CI 2.5–19.68, p 1.70E-04-logit function vs OR 1.75, 95% CI 1.10–2.78, P = 1.82E02).23 This means a greater benefit with T-DM1 in patients with high ERBB2 mRNA expression, in concordance with other studies.22,25,26
Previously, Antolín et al. also found that high levels of ERBB2 gene amplification by fluorescent in situ hybridization (FISH) had a higher pCR in patients treated with trastuzumab and chemotherapy. ERBB2/CEP17 ratio > 5 had a pCR rate of 65%, while those with a ratio ≤ 5 had a pCR rate of 32% (P = .002).27
Recently, Sanfeliu-Torres et al. described the ability of ERBB2 mRNA expression to predict clinical status of HER2-low and zero, with an accuracy of 65.5%.23
HER2DX in HER2+/HR+ BCIn PerELISA trial, patients sensitive to neoadjuvant letrozol in combination with trastuzumab and pertuzumab showed an association between pCR and HER2DX pCR and ERBB2 mRNA scores (P = .008 and P = .003, univariate logistic regression model; ROC [AUC] = 0.803 and 0.896).28 pCR was higher in HER2DX pCR score high vs low (100% vs 7.7%, respectively; P < .004). Similar results were found with pCR and HER2DX ERBB2 mRNA score high vs low (53.3% vs 0.0%, respectively; P = .001).28 In endocrine resistance disease (15%), pCR was 80% and they did not find an association between HER2DX and pCR.28
DiscussionDuring the last years, important improvements in the development of effective anti-HER2 drugs took place. However, not all patients benefit from the same duration and type of treatments. In this context, it is essential to develop genomic tests, with clinicopathological and molecular biomarkers, as HER2DX, in order to improve response, safety, and quality of life with neo/adjuvant therapies.
Clinical characteristics, as tumor size and lymph nodes, remain one of the most important prognostic factors in all BC subtypes.29,30 Tumor stage is an independent predictor of pCR (half of patients with cT3–T4 reached pCR compared to cT1–T2) and lymph node positive disease has substantially worse prognosis in all breast cancer subtypes.29,30
In 2014, Prat et al. published the first report suggesting clinical applicability of gene expression assay.31 A retrospective study with patients from NeOAdjuvant Trastuzumab trial, showed that HER2-E tumors had higher pCR rates after trastuzumab-based chemotherapy compared with non-HER2-E.31 This was also supported by a systematic review published by Schettini et al. where HER2-E subtype was associated with pCR after anti-HER2 therapy with or without chemotherapy and independently of HR status.9 In this context, HER2-E is a consistent biomarker of response after neoadjuvant anti-HER2 regimens and was included in the first HER2DX test developed.22
NEOALTTO and CALGB-40601 trials showed a higher pCR rate in HR negative (HR-) disease compared to HR+, although results of CALGB-40601 were not significant.3,32,33 In this sense, expression levels of ESR1 and progesterone receptor gene are higher in RCB-II and RCB-III tumors than in pCR.29 Nevertheless, 10 years survival follow-up in 2 different trials showed similar outcomes between HR+ and HR- patients.3,34 This group of patients with less response but good prognosis might need less intense treatments.
Immune tumor microenvironment seems particularly relevant in cHER2+ disease, therefore, it is important to measure immune-related information. TILS are the simplest method to measure antitumor immune response, but they give raw information and failed to provide independent prediction of pCR beyond molecular subtypes.35 Based on this, HER2DX included 4 immune-related gene signatures that can predict response from anti-HER2 therapies and add information beyond TILS.35,36
Taking into account this information the multi-variable test HER2DX has been developed. The first version included 17 variables not related to pCR, so a new score with clinical features and 27 genes was developed.13 The 27 genes were grouped on 4 signatures regarding clinical response and survival (Table 1). With this new version, HER2DX can identify patients with different risks of relapse and its association with the likelihood to achieve a pCR.13 pCR rates can be variable depending on the score, between 80% and 90% for HER2DX pCR-high and ≤30% for HER2DX pCR-low, independently of the type of therapy and HR status.20
Gene signatures and prognosis.
| Genetic signature | Genes | Prognostic features |
|---|---|---|
| Immune | CD27, CD79A, HLA-C, IGJ, IGKC, IGL, IGLV3–25, IL2RG, CXC8L, LQX1, NTN3, PIM2, POU2AF1, TNFRSF17 | Better response and survival |
| Tumor cell proliferation | EXO1, ASPM, NEK2 and KIF23 | Better response and lower survival |
| Luminal differentiation | BCL2, DNAJC12, AGR3, AFF3 and ESR1 | Lower response and better survival |
| HER2 amplicon | ERBB2, GRB7, STARD3, TCAP | Associated with response and not with survival |
Dieci et al. identified an increased risk of all-site metastasis in stage III and HER2DX high-risk score, and for brain metastasis in HER2-E and HER2DX high-risk score.18 In this, HER2DX can recognize different risks of relapse and personalize cHER2+ eBC follow-up, focusing efforts on early detection of relapse in high-risk populations.18
The level of ERBB2 mRNA was found to be the best predictor of response to trastuzumab–pertuzumab combination in the absence of chemotherapy.25,28,36 HER2-E/ERBB2 high tumors are the most HER2-addicted and sensitive to anti-HER2 therapies that might benefit from chemotherapy-free strategies.25 These findings led to the development of a validate assay, which can be used for primary endpoints in prospective trials for cHER2+ eBC.13,25
HER2DX results in perELISA trial demonstrated that is a strong predictor of response to endocrine therapy (ET) in combination with trastuzumab and pertuzumab.28 Patients with HER2DX pCR / ERBB2 score high had better response to this treatment, meanwhile, HER2DX pCR low-predicted response to neoadjuvant letrozol monotherapy.28 These results can open a door to different strategies in the future.
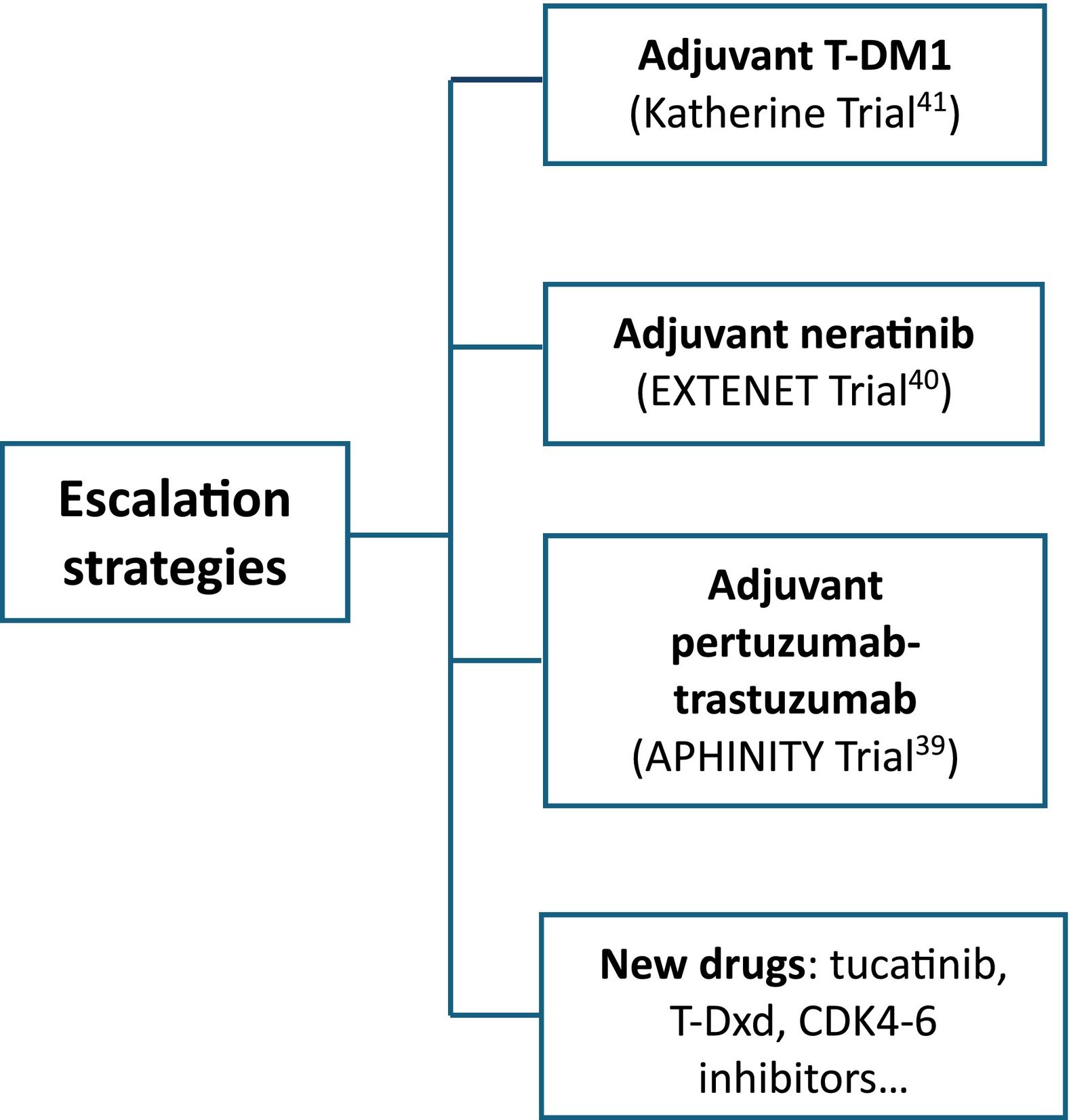
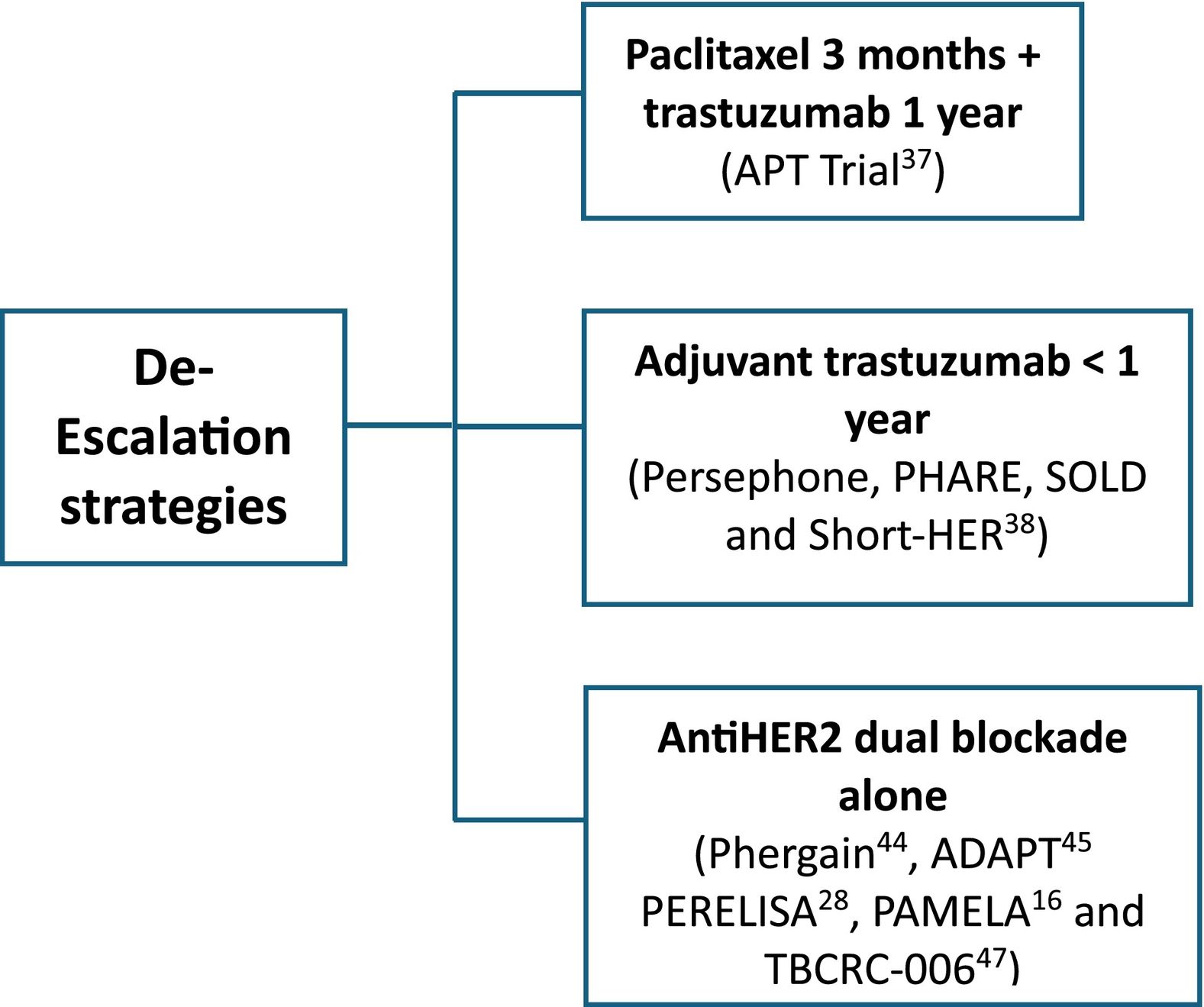
HER2DX is a prognostic score built to identify different populations in cHER2+ eBC who can be candidates for escalation and de-escalation treatments. Various strategies have been evaluated in this sense (Figs. 1 and 2).
In stage I disease with tumor size between 2 and 3 cm, controversy exists with de-escalating adjuvant treatment to paclitaxel 3 months plus trastuzumab 1 year.37 In 10 years analysis of APT trial, HER2DX was associated with DFS and recurrence-free interval and can better identify candidates for this approach.37
Reduction in the duration of adjuvant trastuzumab was tested in 4 clinical trials with conflicting results.38 A metaanaylisis revealed a DFS HR of 1.28 (95% CI 1.09–1.36), supporting longer duration of adjuvant trastuzumab with double cardiac events.38 HER2DX could help to identify patients with low risk of recurrence and cardiac comorbidities, who would be candidates for shorter duration approaches.13
In stage II–III, escalate strategies are based on systemic pertuzumab, neratinib, T-DM1, and new drugs (tucatinib, trastuzumab-deruxtecan (T-Dxd), CDK4–6 inhibitors…) Absolute benefit of adjuvant pertuzumab and neratinib was demonstrated to be low (3% and 5.1%, respectively, in DFS).39,40 In patients who have residual disease, estimated DFS at 3 years is 88.3% for T-DM1 and 77% for trastuzumab.41 HER2DX risk score can help de-escalation strategies identifying patients with low-risk disease regardless pCR and adjuvant T-DM1.20 Despite this, only 16% of oncologist have access to the test in clinical practice.42
Current treatment paradigms involving polychemotherapy and up to 3 anti-HER2 regimens, with 3 year DFS close to 90%, hightlights the need to better define patients groups prognosis in order to avoid unnecessary toxicity and cost.38
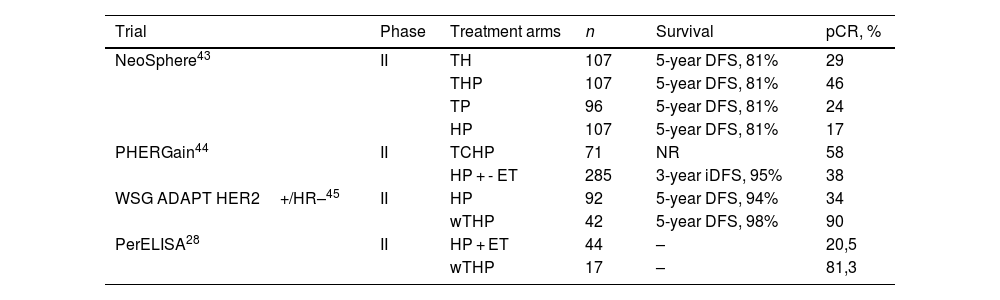
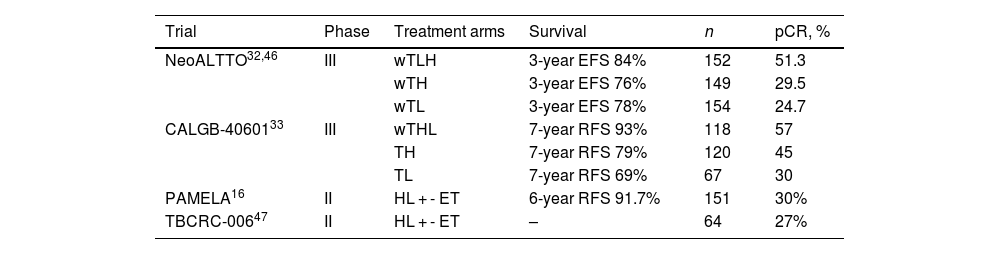
Some tumors are so dependent on HER2 that cure can be achieved with neoadjuvant dual HER2 blockade and allow multi-agent chemotherapy de-escalating38 (Table 2). NeoSphere, PHERGain, WSG ADAPT, and PerELISA trials showed beneficial effects of neoadjuvant pertuzumab and trastuzumab (Table 2). The NeoSphere and PHERGain trials demonstrated high pCR rate with dual blockade in a subgroup of patients.43,44 The WSG ADAPT showed that multi-agent chemotherapy can be safetly omitted in HR-HER2+ eBC.45 In PerELISA trial, patients selected by initial response to letrozol reached pre-specified pCR with trastuzumab and pertuzumab.28 Benefit from trastuzumab and lapatinib was shown in NeoALTTO, CALGB-40601, Pamela, and TBCRC-006 trials (Table 3). Despite the increase in pCR, there has been no improvement in survival and, as the toxicity profile increases, it is not indicated in clinical practice.16,32,33,46,47
Clinical trials with neoadjuvant dual blockade pertuzumab and trastuzumab.
| Trial | Phase | Treatment arms | n | Survival | pCR, % |
|---|---|---|---|---|---|
| NeoSphere43 | II | TH | 107 | 5-year DFS, 81% | 29 |
| THP | 107 | 5-year DFS, 81% | 46 | ||
| TP | 96 | 5-year DFS, 81% | 24 | ||
| HP | 107 | 5-year DFS, 81% | 17 | ||
| PHERGain44 | II | TCHP | 71 | NR | 58 |
| HP + - ET | 285 | 3-year iDFS, 95% | 38 | ||
| WSG ADAPT HER2+/HR–45 | II | HP | 92 | 5-year DFS, 94% | 34 |
| wTHP | 42 | 5-year DFS, 98% | 90 | ||
| PerELISA28 | II | HP + ET | 44 | – | 20,5 |
| wTHP | 17 | – | 81,3 |
Abbreviations: C, carboplatin; ET, endocrine therapy; H, trastuzumab; NR, not reported; P, pertuzumab; pCR, pathological complete response; T, docetaxel; wT, paclitaxel.
Clinical trials with neoadjuvant dual blockade lapatinib and trastuzumab.
| Trial | Phase | Treatment arms | Survival | n | pCR, % |
|---|---|---|---|---|---|
| NeoALTTO32,46 | III | wTLH | 3-year EFS 84% | 152 | 51.3 |
| wTH | 3-year EFS 76% | 149 | 29.5 | ||
| wTL | 3-year EFS 78% | 154 | 24.7 | ||
| CALGB-4060133 | III | wTHL | 7-year RFS 93% | 118 | 57 |
| TH | 7-year RFS 79% | 120 | 45 | ||
| TL | 7-year RFS 69% | 67 | 30 | ||
| PAMELA16 | II | HL + - ET | 6-year RFS 91.7% | 151 | 30% |
| TBCRC-00647 | II | HL + - ET | – | 64 | 27% |
Abbreviations: ET, endocrine therapy; H, trastuzumab; L, lapatinib; pCR, pathological complete response; T, docetaxel; wT, paclitaxel.
Two systematic reviews that compared dual anti-HER2 blockade vs monotherapy revealed an increase in pCR with dual antibody.48,49 Zhang et al. did not find an increment in cardiac toxicity, however, grade 3–4 toxicities were most frequent with combined anti-HER2 therapy.48 On the other hand, Wang et al. did not observe an increase in serious adverse events and cardiotoxicity, but neither demonstrated a difference in DFS.49
HER2DX will guide the use of systemic therapy in HER2+ eBC, but at the moment, we only have retrospective studies (Table 4) and further prospective validation is need to study escalation or de-escalation options.12,50 In this context, the DEFINITIVE project pretends to demonstrate that the use of HER2DX assay improves quality of life and maintains drug efficacy in HER2+ eBC across 44 hospitals worldwide.51
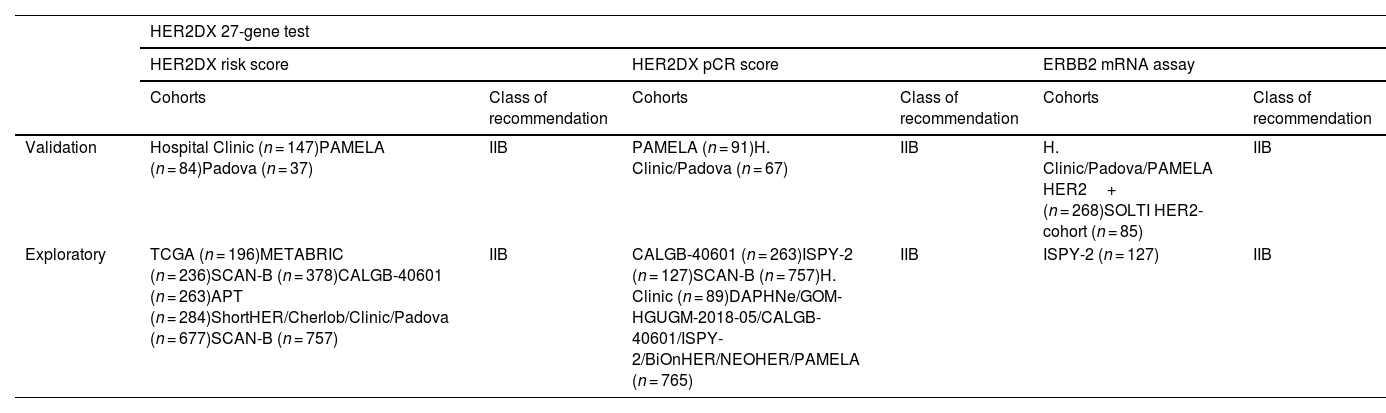
Cohorts of patients where HER2DX was retrospectively validated and class of recommendation across levels of evidence:
| HER2DX 27-gene test | ||||||
|---|---|---|---|---|---|---|
| HER2DX risk score | HER2DX pCR score | ERBB2 mRNA assay | ||||
| Cohorts | Class of recommendation | Cohorts | Class of recommendation | Cohorts | Class of recommendation | |
| Validation | Hospital Clinic (n = 147)PAMELA (n = 84)Padova (n = 37) | IIB | PAMELA (n = 91)H. Clinic/Padova (n = 67) | IIB | H. Clinic/Padova/PAMELA HER2+ (n = 268)SOLTI HER2-cohort (n = 85) | IIB |
| Exploratory | TCGA (n = 196)METABRIC (n = 236)SCAN-B (n = 378)CALGB-40601 (n = 263)APT (n = 284)ShortHER/Cherlob/Clinic/Padova (n = 677)SCAN-B (n = 757) | IIB | CALGB-40601 (n = 263)ISPY-2 (n = 127)SCAN-B (n = 757)H. Clinic (n = 89)DAPHNe/GOM-HGUGM-2018-05/CALGB-40601/ISPY-2/BiOnHER/NEOHER/PAMELA (n = 765) | IIB | ISPY-2 (n = 127) | IIB |
Class of recommendation IIB: II: Evidence from ≥1 well-designed clinical trial, without randomization; from cohort or case-controlled analytic studies (preferably from >1 center); from multiple time series; or from dramatic results from uncontrolled experiments. B: Moderate evidence to support a recommendation for use.
A pilot study of clinical applicability of HER2DX test revealed a change in treatment decisions related to the test in 56% of cases.21
ConclusionThe development of new therapies has generated the need of biomarkers to outline personalized therapies, with the aim to improve survival and quality of life. In this direction, HER2DX assay was developed, with 2 independent scores, that predict long-term prognosis and the likelihood to achieve a pCR to neoadjuvant therapy. The studies carried out with HER2DX provided a hope to cover the need of validate biomarkers in patients with HER2+ eBC, but further prospective analyses are required to confirm this results.
FundingThis review did not receive financial support.
Ethical considerationsThe authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Author contributionsAll authors contribute equality to conception and design, collection of data, manuscript writing and final approval of manuscript.
Declaration of competing interestAuthors have no conflicts of interest to declare.
Breast cancer unit of UniversityHospital La Coruña and Drug Development Department Gustave Roussy institute.