This work describes the genetic characterization of Cryptosporidium and Giardia involved in an outbreak in a nursery school in Granada, Spain, that affected seven children under the age of 4.
MethodsNucleic acids were extracted from the seven stool samples positive to Cryptosporidium or Giardia by microscopy and/or immunochromatography. The species and subtypes of Cryptosporidium were identified by PCR-RFLP and PCR of the SSUrRNA and gp60 genes, respectively. The assemblages of Giardia duodenalis isolates were characterized by PCR of the tpi gene. PCR products were sequenced and analyzed.
ResultsAll of the isolates were positive for Cryptosporidium hominis. Five of them belonged to subtype IaA11R2, one to subtype IbA10G2R2, and the other could not be identified. Three of these samples were positive for G. duodenalis by PCR, two belonging to the assemblage A, and the other one to assemblage B.
DiscussionThis is the first report of Cryptosporidium hominis subtype IaA11R2 as a cause of an outbreak in Europe where subtype IbA10G2R2 is the most frequently identified. In the case of Giardia, an outbreak could not be confirmed because of the low number of positive samples and the low genetic variability of the amplified fragments for assemblage A of tpi gene.
ConclusionsA new subtype, of Cryptosporidium hominis named IaA11R2, has been described as a cause of an outbreak in a nursery school in Granada, Spain. However an outbreak of giardiasis could not be confirmed.
Este trabajo describe la caracterización genética de Cryptosporidium y Giardia implicados en un brote en una guardería de Granada, España, que afectó a 7 niños menores de 4 años de edad.
MétodosLa extracción de ácidos nucleicos se realizó a partir de las 7 muestras de heces positivas para Cryptosporidium o Giardia mediante técnicas de microscopia y/o inmunocromatografía. Las especies y subtipos de Cryptosporidium fueron identificados por PCR-RFLP y PCR de los genes SSUrRNA y gp60, respectivamente. Los genotipos de los aislamientos Giardia duodenalis fueron caracterizados por PCR del gen tpi. Los productos de PCR obtenidos fueron secuenciados y analizados.
ResultadosTodos los aislamientos fueron positivos para Cryptosporidium hominis. Cinco de ellos correspondieron al subtipo IaA11R2, uno al subtipo IbA10G2R2 y otro no pudo ser identificado. Tres de las muestras resultaron positivas para G. duodenalis por PCR, 2 de ellas pertenecientes al genotipo A y la restante al genotipo B.
DiscusiónEsta es la primera identificación del subtipo IaA11R2 de Cryptosporidium hominis como causa de un brote en Europa, donde el subtipo IbA10G2R2 es el más frecuentemente identificado. En el caso de Giardia, el bajo número de muestras positivas y la baja variabilidad genética del fragmento del gen tpi amplificado para el genotipo A no permitieron confirmar un brote.
ConclusionesUn nuevo subtipo de Cryptosporidium hominis, denominado IaA11R2, ha sido descrito como causa de un brote en una guardería de Granada, España. Sin embargo, los resultados obtenidos no permiten confirmar un brote de giardiasis.
Cryptosporidium spp. (Protoctista, Apicomplexa) is a protozoan parasite that causes gastrointestinal disorders in a large number of hosts including humans and animals. Twenty-six species and more than 40 genotypes have been identified to date, but Cryptosporidium hominis and Cryptosporidium parvum are the two species that cause 90% of all infections in immunocompetent and immunosuppressed individuals.1,2 Both species have been found in Spain in waters, in bivalve molluscs and humans. The predominant species in animals is C. parvum whereas in humans, the highest prevalence of C. hominis has been documented. Nevertheless, it seems that the predominant species depend on the rural or urban environment, with C. parvum more commonly detected in rural and C. hominis in urban areas.3,4 Both species have been reported to cause outbreaks in the general population and in children under 2 years of age. However, since cryptosporidiosis in Spain is not a notifiable disease, data that do exist concerning this disease are scarce and are not based upon 100% of the population. The discovery of some species such as Cryptosporidium meleagridis, Cryptosporidium felis or Cryptosporidium ubiquitum in Spain, and its importance in human epidemiology has also been documented.4,5 The most recent method used for identifying subtype families and subtypes of Cryptosporidium spp. is the one based on the analysis of the partial sequence of the gene coding for a polymorphic glycoprotein expressed on the sporozoite surface (GP60). To the date there are several subtype families described: nine belonging to C. hominis species (Ia–Ig), 14 to C. parvum (IIa–IIl), seven to C. meleagridis (IIIa–IIIf) and six to Cryptosporidium fayeri (IVa–IVf).2,6,7 Each subtype group is subdivided into subgenotypes based on the number of tri-nucleotide repeats coding for the amino acid serine and named following the rules described by Sulaiman et al.8,9
There are few studies about Cryptosporidium subtypes in Spain but in general, the predominant subtype is the IbA10G2R2 subtype, although others such as the IaA18R3 subtype has been identified in Navarra in the summer of 2012.10
In addition, human giardiasis is not a compulsory notifiable disease in Spain, so its spread is difficult to assess. A total of 16974 cases of Giardia duodenalis infections were notified to the microbiological information system between 1989 and 2010. Children under 5 years of age accounted for 42.7% of reported cases.11 Most reported outbreaks involved person-to-person contact, although waterborne transmission was also documented. Eight assemblages of G. duodenalis have been described to date, but typically only assemblages A and B are associated with human infections. Subassemblage AI mostly infects animals whereas humans are mostly infected with subassemblage AII. In Spain, assemblage B is the most prevalent in human population, and also subassemblage AII and a mixed infection of subassemblage AII and assemblage B have been identified. Subassemblage AI has been found in cattle and sheep and it has not been reported so far in humans.11,12
The aim of this study was the genetic characterization of Cryptosporidium spp. and G. duodenalis involved in an outbreak in a nursery school in Granada, Spain.
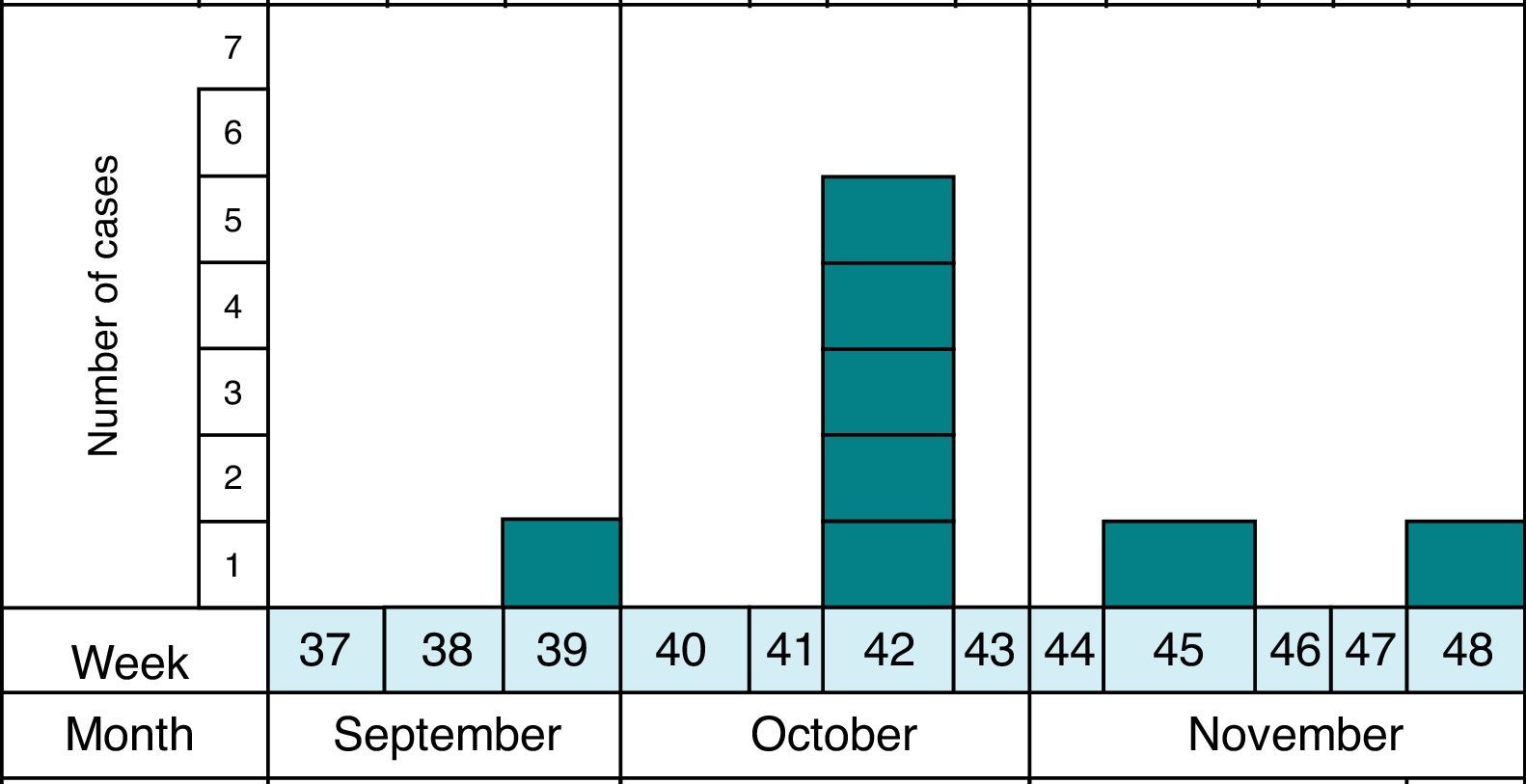
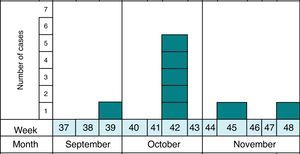
MethodsAs previously reported,13 the notification and confirmation of a suspected outbreak of acute gastroenteritis (AGE) in a nursery in the municipality of Maracena (Granada, Spain) occurred between September and December 2013. Cases were defined as subjects with diarrhea (defined as the occurrence of at least two consecutive unformed stools) or abdominal pain as clinical criteria and stool samples positive for G. duodenalis or Cryptosporidium by microscopy and/or immunochromatography as laboratory criteria. In addition, the definition of suspect (every person who meets the clinical criteria, have epidemiological relationship and has been reported by epidemiological survey), probable (every person who meets the clinical criteria, have epidemiological relationship and has been reported by searching through digital medical records) and confirmed case (every person who meets the clinical and laboratory criteria) was made. The methodology to identify the causal agent and its possible domestic origin was created. Fig. 1 shows the distribution of all cases of cryptosporidiosis occurring in Granada in the study period. At all times the outbreak has been contextualized with confirmed cases.
During the epidemic period, 70 children between 0 and 3 years old went to the nursery, which had five classes on two floors and five carers permanently assigned to class. Most of the families of affected children belong to low-medium socio-economic status.
Eighty-two stool samples from 75 subjects were collected and analyzed, including 60 children under the age of 6 who had direct contact at the nursery, nine persons aged between 6 and 16 years old as family members, and six nursery workers older than 16 years old.13 Seven individuals were sampled twice.
In this previous study and in order to rule out causes of infection, routine stool culture and antigen detection of rotavirus and adenovirus were performed for all samples. In addition, the presence of Cryptosporidium spp. and G. duodenalis was investigated by immunochromatography (IC) (CerTest Crypto/Giardia, Certest Biotec S.L., Zaragoza, Spain) and in all cases the stool samples were analyzed for the presence of other intestinal parasites by microscopy following standard formalin–ethyl acetate concentration. Also, smears from the concentrated samples were stained with a modified Ziehl–Neelsen stain and examined by means of microscopy to detect Cryptosporidium oocysts. In the previous study, all samples were found to be negative on bacteriological isolation and detection of viral antigens. In the parasitological study, seven (13.3%) of the samples from children were positive to Cryptosporidium by microscopy and immunochromatography and three of them were also positive for G. duodenalis (5%), and one for Blastocystis hominis. Adults were not parasitized. None of the parasitized children had a history of previous traveling abroad.13 These seven samples that tested positive for microscopy and immunochromatography techniques were assessed by molecular methods in this study.
All samples positive to Cryptosporidium and or G. duodenalis (7) by microscopy or IC were analyzed by molecular biology techniques to genetic characterize both Cryptosporidium and G. duodenalis isolates.
Nucleic acids were extracted using DNA stool kit (Norgen Biotek Corp., Ontario, Canada) following the manufacturer's instructions and stored at −20°C until processing. A 826–864bp fragment of 18S rRNA subunit (SSUrRNA) was amplified by PCR using the primers and conditions described by Xiao et al. (1999) and restriction fragment polymorphism (PCR-RFLP) with SspI and VspI (Promega, Madison, USA) was performed to identify Cryptosporidium species.14 SSU1 and SSU2 primers were used in the first PCR in 25μl of reaction mix with the following conditions: 10× PCR buffer (Biotools B & M Labs, S.A., Madrid, Spain), 2mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200μM, each primer at a concentration of 200nM, 2.5U of Taq polymerase, and 2μl of DNA template. A total of 35 cycles, consisting of 94°C for 45s, 55°C for 45s, and 72°C for 1min, were performed; an initial hot start at 94°C for 3min and a final extension step at 72°C for 7min were also included. For secondary PCR, 2μl of the primary PCR product were used, and the reaction was performed with SSU3 and SSU4 primers with identical PCR mixture and cycling conditions used in the primary PCR, except 1.5mM MgCl2 was used in a 50μl PCR mixture of final volume. To identify the families and subtypes, a fragment of the gp60 gene was amplified using nested PCR.15 The fragment of 400bp was amplified using primers AL3531, AL3533 in primary PCR and AL3532, LX0029 in the secondary PCR.16 Both reactions were performed with a mixture containing 10× PCR buffer (Biotools B & M Labs, S.A., Madrid, Spain), 1.5mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200μM, each primer at a concentration of 200nM, 2.5U of Taq polymerase, and 5μl of DNA template or primary PCR product in a final volume of 50μl. Cycling program include an initial hot start at 95°C for 3min, 40 cycles, with 95°C for 45s, 52°C for 45s, and 72°C for 1min; and a final extension step at 72°C for 10min.
G. duodenalis assemblages were determined by a PCR of a 530bp fragment of triosephosphate isomerase (tpi) gene.16 AL3543 and AL3546 primers were used in a primary PCR with a reaction mixture containing 10× PCR buffer (Biotools B & M Labs, S.A., Madrid, Spain), 3mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200μM, each primer at a concentration of 200nM, 2.5U of Taq polymerase, and 5μl of DNA template in a final volume of 50μl. A secondary PCR was performed using AL3543 and AL3546 primers at a concentration of 200nM and 100nM respectively, in 10× PCR buffer (Biotools B & M Labs, S.A., Madrid, Spain), 2mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200μM, 2.5U of Taq polymerase, and 5μl of primary PCR product in a final volume of 50μl. Both reactions were performed under the following conditions: a hot start of 94°C for 5min, 35 cycles, with 94°C for 45s, 50°C for 45s, and 72°C for 1min; and a final extension step at 72°C for 10min.
In all reactions positive and negative controls have been included with DNA of a previous characterized isolate and replacing the DNA by sterile double distilled water, respectively.
All PCR products were purified with Illustra™ GFX™ PCR DNA Gel Band Purification Kit (GE Healthcare Life Sciences, Bukinghamshire, UK) and direct sequenced in both directions in an external laboratory (Citogen, S.L., Zaragoza, Spain). The nucleotide sequences obtained were analyzed and compared with those registered in GenBank using Chromas 2.4.1.0, BioEdit 7.0.0 (http://www.mbio.ncsu.edu/bioedit/bioedit) and MEGA 6 (http://www.megasoftware.net/mega.php).
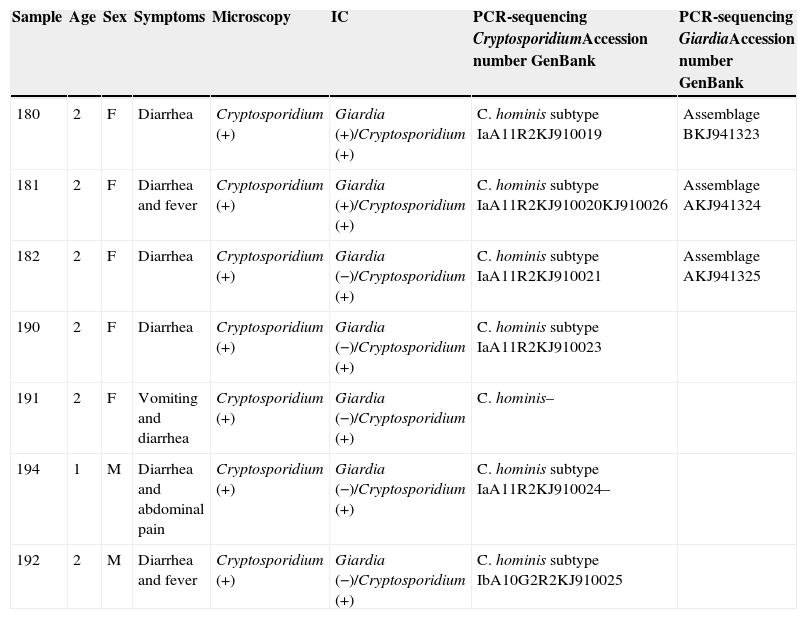
The DNA sequences obtained have been deposited in the genetic sequence database (GenBank) at the National Center for Biotechnology Information (Table 1).
Resume of clinical and epidemiological data with laboratory results.
| Sample | Age | Sex | Symptoms | Microscopy | IC | PCR-sequencing CryptosporidiumAccession number GenBank | PCR-sequencing GiardiaAccession number GenBank |
|---|---|---|---|---|---|---|---|
| 180 | 2 | F | Diarrhea | Cryptosporidium (+) | Giardia (+)/Cryptosporidium (+) | C. hominis subtype IaA11R2KJ910019 | Assemblage BKJ941323 |
| 181 | 2 | F | Diarrhea and fever | Cryptosporidium (+) | Giardia (+)/Cryptosporidium (+) | C. hominis subtype IaA11R2KJ910020KJ910026 | Assemblage AKJ941324 |
| 182 | 2 | F | Diarrhea | Cryptosporidium (+) | Giardia (−)/Cryptosporidium (+) | C. hominis subtype IaA11R2KJ910021 | Assemblage AKJ941325 |
| 190 | 2 | F | Diarrhea | Cryptosporidium (+) | Giardia (−)/Cryptosporidium (+) | C. hominis subtype IaA11R2KJ910023 | |
| 191 | 2 | F | Vomiting and diarrhea | Cryptosporidium (+) | Giardia (−)/Cryptosporidium (+) | C. hominis– | |
| 194 | 1 | M | Diarrhea and abdominal pain | Cryptosporidium (+) | Giardia (−)/Cryptosporidium (+) | C. hominis subtype IaA11R2KJ910024– | |
| 192 | 2 | M | Diarrhea and fever | Cryptosporidium (+) | Giardia (−)/Cryptosporidium (+) | C. hominis subtype IbA10G2R2KJ910025 |
IC, immunochromatography.
The diagnosis of these seven positive samples was confirmed with the nested PCR analysis of the SSUrRNA PCR gene. RFLP analysis of the secondary PCR products with restriction enzymes SspI and VspI showed a band profile consistent with C. hominis patterns: 450, 267, 108-bp and 561, 111-bp, respectively.
When PCR of gp60 gene was performed to determine Cryptosporidium subtype, the expected amplicon of about 400bp was obtained for the seven samples. The sequence analysis showed that five isolates belong to subtype IaA11R2, one to subtype IbA10G2R2 and the other one could not be sequenced. The sequences obtained for the amplification of the fragment of the SSUrRNA gene were identical to each other, while those corresponding to the amplified fragment of the gene encoding the glycoprotein GP60 for the subtype IaA11R2 presented homologies between 99.4% and 100%. The single nucleotide polymorphisms (SNP) found in these sequences are shown in Fig. 2. None of them affects the variable tandem repeat region of the serine-coding trinucleotide (TCA/TCG/TCT), which is the primary target of the gp60 typing methods or the repetitions located immediately after the trinucleotide repeats and represented by the letter R. Changes in nucleotide 227 (C→A) in the nucleotide sequence obtained for the strain 194 implies changing S by Y in position 85 of the resulting protein. In the isolate 182, a change of C to T at positions 305 and 308 of the nucleotide sequence obtained for the amplified fragment involves a change of T to I at positions 102 and 103 of the amino-acidic sequence of the protein.
On the other hand three of the children's samples, all of them containing Cryptosporidium subtype IaA11R2, were also positive for G. duodenalis by PCR, identifying the assemblages A and B of G. duodenalis in two and one sample, respectively (Fig. 2).
Table 1 shows a resume of clinical and epidemiological data with laboratory results and accession numbers in GenBank for the sequences obtained in this work.
DiscussionC. hominis has been identified as the cause of an outbreak that affects seven children in a nursery in Granada (Spain). Human cryptosporidiosis is commonly caused by C. hominis or C. parvum, being the transmission of C. hominis exclusively anthroponotic.17
In this work, two subtypes have been identified in six of the seven cases, a new non-reported subtype named IaA11R2, found in five of the six cases, and subtype IbA10G2R2 the other case. The coincidence in species and subtype, along with the high homology of the sequences obtained for the five cases in which the subtype found was IaA11R2 and epidemiological evidence supports the occurrence of an outbreak of cryptosporidiosis.
In Spain, few outbreaks have been described in the literature. In 2000, Rodriguez-Salinas Pérez et al. described an outbreak in Guadarrama (Madrid),18 which affected 21 preschool children; in 2003, Pedalino et al. reported an outbreak which affected 391 people with cryptosporidiosis among tourists who stayed in the same hotel in Mallorca.19 Subsequently, in 2012, Artieda et al. reported an outbreak that affected 26 persons in a child day-care center in Guipuzcoa.20 However, the determination of Cryptosporidium species and subtype was not performed in any of these cases, inferring the presence of an outbreak based on epidemiological data.
As predominant species, C. hominis, C. parvum, C. meleagridis, C. felis and C. ubiquitum have been found infecting humans in Spain, being C. hominis predominant in urban environments, but also with a high presence in rural environments.4,5,21
Cryptosporidium families have geographical distribution, and in Europe and Australia most C. hominis infections are caused by the Ib family.22 Of the C. hominis subtypes, the Ia subtype has been identified in humans in some countries such as Peru,23 China,24 Australia,25 Malaysia,26 Bangladesh,6 USA,27 although in no case has it been the most common subtype. As far as we know, in Europe, it has been detected only in Portugal and recently in Spain.10,28 Also, Ia family has been associated with diarrhea, nausea and vomiting in children.29
With respect to subtypes of C. hominis, the identification of IbA10G2R2 and IaA21G1R1 has been reported in a study conducted by Jex and Gaser (2008) in people who have traveled to Spain.25 These authors suggest that the latter subtype, IbA10G2R2, is the main subtype of C. hominis in Europe. Also, Abal-Fabeiro et al. (2014) identified the family Ib as the subtype of C. hominis predominant in Santiago de Compostela.21 So far as we know, this paper describes for the first time the subtype IaA11R2 as the cause of an outbreak in a nursery. The literature up to now shows that subtype IaA11R3, which is the most similar to IaA11R2 was found in Bangladesh,6 IaA11R4 in Peru,23 IaA19R3 in Portugal28 and in Canada,30 IaA17R1 in Australia31 and IaA12R3 in USA (but in that case the patient had traveled to Pakistan).31
However, the presence of G. duodenalis in the sample from three of the children concerned is interesting. In addition, G. duodenalis isolates belong to different G. duodenalis assemblages, two of them to G. duodenalis assemblage A and one to G. duodenalis assemblage B. The homology between the sequences of G. duodenalis assemblage A was 99.7%, although the fragment studied did not exhibit enough polymorphism to differentiate within the G. duodenalis assemblage A isolates. This low variability coupled with the small number of cases of the same assemblage prevents the differentiation of isolates. Therefore the presence of a simultaneous outbreak of cryptosporidiosis and giardiasis could not be assured. Several studies in different regions of Spain have detected G. duodenalis assemblage A, although G. duodenalis assemblage B was always identified as prevalent.12,32–34G. duodenalis has been detected only in three of the children together with cryptosporidiosis and of them two belonged to assemblage A. Both parasites could be acquired together but this fact could not be confirmed. G. duodenalis assemblage A was also considered to be human-specific, although two studies have reported its presence in dog samples.35,36 Also this assemblage has been associated with the presence of symptoms in children while assemblage B has been associated with asymptomatic infection.33 In this case asymptomatic individuals have not been detected. Furthermore, the simultaneous presence of Cryptosporidium and G. duodenalis in various patients did not allow to establish a relationship between symptoms and assemblage of G. duodenalis, in contrast with those reported by Mateo et al. (2014), who in a study conducted in nurseries in Madrid, obtained 82.4% of positive cases for G. duodenalis and Cryptosporidium asymptomatic, with all the G. duodenalis identified belonging to subassemblage BIV.34
Our results indicate that more studies about Cryptosporidium and G. duodenalis genotyping are needed to identify the populations that are spreading in Spain, and establish their potential geographical specificity and their transmission path.
Ethical statementThe manuscript does not contain clinical studies or patient data.
Conflict of interestThe authors declare that they have no conflict of interest.
This work was funded by DGA-FSE Research Team B82, and Project UZ2013-FIS-02 University of Zaragoza.