The differences in the expression profiles of colonic miRNAs between β-lactoglobulin (β-Lg) allergic mice and normal mice were analyzed to investigate the important role of the miRNA regulation mechanism in the pathogenesis of cow’s milk allergy.
MethodsThe present study performed Illumina sequencing to characterize the miRNA profile changes in mouse colon responding to β-Lg challenge. Target genes were predicted by TargetScan 50 and miRanda 3.3a algorithms and assessed by GO and KEGG analysis. The expression levels of selected miRNAs and cytokine production were verified by cell transfection and quantitative RT-PCR.
ResultsA total of 15 miRNAs were diversely expressed between the colon of the normal and β-Lg-sensitized mice (P < 0.05, fold change of >1.50 or <0.67), including six up-regulated miRNAs and nine down-regulated miRNAs, among which seven miRNAs were validated using qRT-PCR. GO enrichment and KEGG pathway analyses further revealed that biological process, protein binding, cytoplasm and the pathways of cancer were significantly enriched, which were closely connected to the allergic inflammation development. Additionally, six key functional interaction pairs in β-Lg allergy were identified in miRNA prediction algorithms and verified using qRT-PCR.
ConclusionsWe can conclude that our results suggested that the miRNAs regulation network participated in the pathogenesis of cow’s milk allergy.
Cow’s milk allergy (CMA) with β-lactoglobulin (β-Lg) represents a major allergen protein that is one of the most common food hypersensitivities, which could cause a variety of skin, gastrointestinal or respiratory tract disorders in 2–3% infants and young children. The existence and severity of CMA have been strongly linked to dysregulated immune responses, including IgE-mediated mast cells/eosinophils activation, abnormal CD4+ lymphocytes subsets differentiation and excessive proinflammatory cytokine production, etc.1 In addition to Th2-dominant immune imbalances, growing evidence supports that augmented Th17 cells and diminished regulatory T cells (Treg) were also involved in the pathogenesis of allergic disease.2 Moreover, intracellular signaling pathways as key regulators of immunologic processes contributed to Th cell differentiation. For example, the JAK-STAT pathway has been reported to promote Th2-mediated inflammation in allergic rhinitis3 and severe asthma,4 as well as to trigger a predominantly Th17 response in autoimmune arthritis mice.5 The NF-κB signal was believed to be a key prerequisite for Th cell activation, and defects in Th26 and Th17 recruitment were found in NF-κB-lack mice.7 An activated TGF-β pathway, such as TGF-beta 1 signaling, has been shown to be closely associated with Treg expansion in asthmatic mice, and resulted in promoting and sustaining tolerance to allergens.8 Although the above studies have characterized some potential therapeutic targets for preventing CMA, they failed to provide evidence about the related regulatory mechanism at post-transcriptional level.
MiRNAs are newly discovered non-coding RNAs that are 21–25 nucleotides long, which rapidly and stably modify post-transcriptional gene expression in various biological processes. Increasing studies focused on the regulation of miRNAs and their target genes on the innate and adaptive immune process, including lymphocyte activation and autoimmune.9 Some unique miRNAs have been described as potential biomarkers in allergic diseases. For instance, Guan-Nan Tang et al. found that asthma mouse exhibited the aberrant miRNAs profile and 14 miRNAs were differently expressed between the normal and asthma samples.10 Maximilian et al. revealed that 29 specific miRNAs were remarkably changed in the lungs from the murine ovalbumin-induced allergic airways model.11 However, little is known about the altering miRNA expression pattern during CMA development. Furthermore, recent work suggested the capacity of miRNAs on mediating T cell proliferation and differentiation. On the one hand, miR-181a, miR-146a and miR-146b were reported to have a positive correlation with increased Th2 numbers in murine models with acute asthma.12 Moreover, miRNA-21 could increase Th17 levels in an allergic murine model.13 Clotaire et al. suggested that miR-19b-3p directly targeted on JAK/STAT, and up-regulated miR-19b-3p might be responsible for cell self-renewal in colorectal mucosa of colorectal cancer patients.14 On the other hand, over-expressed miR-15b and miR-16 were observed to enhance Treg percentages by suppressing HIF-2 in murine naive T cell.15 FOXP3, which was a master regulator of Treg, could be induced by down-regulated miR-34a expression in allergic asthma mice.16 Therefore, studies on the regulating function of miRNA would provide a new insight for the pathogenesis of CMA.
In light of the above, this study aimed to investigate the role of miRNAs in response to β-Lg allergy. The miRNA-microarray and corresponding target genes were explored by Illumina sequencing and bioinformatics analysis, respectively. Subsequently, allergy-related biological processes and signaling pathways driven by miRNA-target interactions were further defined to explain the associated molecular regulation. These data will help to better understand the molecular mechanisms of CMA pathogenesis.
Materials and methodsβ-Lg-sensitized murine model and sample obtainingAs described in our previous study,17 five to sxi-week-old female BALB/c mice (Harbin Veterinary Research Institute, China) were randomly divided into normal group (N) and β-Lg allergic group (A) (n = 15 per group). From day 1 to day 21, the sensitized mice were weekly intraperitoneally injected with 0.2 mL 1 mg/mL β-Lg (Sigma, USA) dissolved in Freund’s adjuvant (Sigma, USA), and the mice treated with sterile saline were designated as the control group. On day 28, allergic mice were orally gavaged with 20 mg β-Lg twice. Two hours after treatment, we collected blood samples from mice by removing their eyes, then all the mice were sacrificed by cervical dislocation and the colon tissue samples were taken and stored in liquid nitrogen for subsequent miRNA analysis. All in vivo manipulations were performed in accordance with the guidelines for Care and Use of Laboratory Animals of Northeast Agricultural University (Permeation number: NEAU-[2016]-9).
Assessment of the hypersensitivity symptomsThe body weight and diarrhea graded of allergic mice were monitored weekly during β-Lg challenge. Diarrhea symptoms were scored as previously described.18 After the final oral β-Lg challenge, the sera levels of mouse mast cell proteasem-1 (MCP-1), β-Lg-specific IgE, interleukin-4 (IL-4), IL-6, IL-13 and IL-17 were measured using specific mouse ELISA kits according to the manufacturer’s instructions (Jiancheng, Nanjing, China). The blood eosinophils were examined with hemocytometer counted under optical microscope staining with May-Grünewald-Giemsa (Solarbio, China). The histological evaluation of the colon was detected by using hematoxylin and eosin (H&E) stain (Solarbio, China).
miRNA library construction and sequencingTotal RNA samples were extracted and harvested from three allergic mice colons (A1, A2, A3) and three control colons (N1, N2, N3) using Trizol reagent (Invitrogen, CA, USA). The quantity and purity of extracted RNA were analyzed by Bioanalyzer 2100 (Agilent, CA, USA) with RIN number >7.0. Total RNA from individual colon was used to prepare a small RNA library according to the protocol of TruSeq small RNA sample prep kit (Illumina, San Diego, USA). Six sequencing libraries were performed by single-end sequencing (36bp or 50bp) using an Illumina Hiseq 2500 at Lianchuan Biological Technology Co., Ltd. (Hangzhou, China).
High-throughput sequence data processing and statistical analysisBriefly, the raw sequence reads obtained from Hiseq were processed by removing the adapter dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, the clean sequence reads at length in 18∼26 nucleotide were mapped to specific species precursors in miRBase 21.0 by BLAST search against mammalian miRNA precursors and mature miRNAs to identify the known miRNAs. The unmapped sequences were predicted by BLASTing against the specific genomes in the miRBase (http://www.mirbase.org/) and predicating the hairpin RNA structures using RNAfold software with default parameters.
To find out the altered miRNAs between the β-Lg allergic mice and the normal, the miRNAs from six small RNA libraries were normalized by obtaining the expression of transcripts per million (TPM) with the following criteria: firstly, normalized expression = actual miRNA count /total count of clean reads*1,000,000. Then, fold change (fold change = log2 (allergic TPM/control TPM)) and Student t-test were used to pinpoint the miRNA expression. The significance threshold was set to be fold change >0.67 and <1.5, and P-value <0.05.
Target gene prediction and functional enrichment analysisTwo computational target prediction algorithms (TargetScan 50 and miRanda 3.3a) were used to identify miRNA binding sites for predicting target genes of differential miRNAs. The data predicted by both algorithms was combined and maintained for the subsequent analysis.
Potential functions and pathways of the predicted target genes were assessed by Gene Ontology (GO) (http://www.geneontology.org/) and Kyoto Encyclopedia Genes and Genomes (KEGG) pathway (http://www.genome.jp/kegg/). The significant criterion of GO terms and KEGG pathways was P -value <0.05.
Validation of miRNAs and key candidate genesTo evaluate the credibility of expression profiling for the miRNAs and their target genes, the differential expressions of seven miRNAs and four mRNAs were identified by quantitative RT-PCR (qRT-PCR) analysis (SYBR fluorescent dye method). U6 and β-actin were used as the internal reference, respectively. The total RNA was extracted from the mice colon tissue using the TRNzol Reagent (Tiangen, Beijing, China) according to the manufacturer’s instructions. In brief, the reverse transcription reaction was performed according to the FastQuant RT Kit (Tiangen, Beijing, China). The qRT-PCR analysis for miRNA and mRNA expression was performed on cDNA in triplicate with the SYBR Green PCR Master PreMix Plus (Tiangen, Beijing, China). Primers for miRNA amplification were designed by stem-loop and listed in Table 1. The miRNA and mRNA relative expression were then quantified using the 2−ΔΔCt method.
Quantitative RT-PCR primers for miRNAs validation.
| Molecule | Sequence (5′-3′) | |
|---|---|---|
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
| mmu-miR-224-5p | Forward | GAGAAACAAGAAGCGCTGTTTA |
| Reverse | ATAGAGTCATTGTCCATAGGCG | |
| mmu-miR-365-2-5p | Forward | CAGTTGCAGCGTTTGACATTAT |
| Reverse | ACTGTAACAGGTGTGGTAACTT | |
| mmu-miR-30a-5p | Forward | CAAGGTGAATGACTTATGCTGG |
| Reverse | CCATACCGTCTGAAGTAATCGA | |
| mmu-miR-504-5p | Forward | GCGAGACCCTGGTCTGCAC |
| Reverse | AGTGCAGGGTCCGAGGTATT | |
| mmu-miR-150-5p | Forward | CCTGTCGCATTGCATGTTAATA |
| Reverse | GGTCTTCGTCGTTATAGTGTCT | |
| mmu-miR-19a-5p | Forward | CAGTACCTCACACGTTTGTCA |
| Reverse | GGAGCCTTTCATGAAATATGGC | |
| mmu-miR-106a-5p | Forward | GAAGGATGATGAAGTTGCCATC |
| Reverse | CTGTTTGATGCTGGTTCCATAC | |
| β-actin | Forward | CGGAATGACGTGAATGCGAAT |
| Reverse | GGCCTGAGATATCCGTCCGC | |
| Tgfβr2 | Forward | GACCTCAAGAGCTCTAACATCC |
| Reverse | GTCATCCACAGACAGAGTAGG | |
| PTEN | Forward | GACCCGTGGCACTGCTGTTTC |
| Reverse | GCTGGCAGACCACAAACTGAGG | |
| SOCS1 | Forward | GAGACCTTCGACTGCCTTTTC |
| Reverse | GGAAGGAACTCAGGTAGTCAC | |
| FOXP3 | Forward | TTTCACCTATGCCACCCTTATC |
| Reverse | CATGCGAGTAAACCAATGGTAG |
RAW264.7 cells were acquired from the cell bank of Shanghai Institute of Cell Biology (Shanghai, China), and cultivated in DMEM complete medium with 10% fetal bovine serum (FBS) at 37 ℃ in a 5% CO2 atmosphere. FBS and DMEM-high glucose medium were obtained from Wisent Inc. (Nanjing, China). The RAW264.7 cells were seeded in a 12-well plate (2 × 105 per well) and preincubated for 24 h. Subsequently, cells in the β-Lg allergic group were incubated for 48 h with β-Lg (1 mg/mL). To assess the effect of miR-19a mimic and inhibitor on β-Lg-induced allergic inflammation, cells were transfected with 60 nM miR-19a mimics or inhibitor using RFect transfection reagent for 36 h.
Statistical analysisExperimental data were expressed as the mean ± standard deviation (SD). Statistical analyses between groups were performed by means of ANOVA. SPSS version 19 (SPSS Inc, Chicago, USA) was used for the analysis. The significant criterion was considered as P -value <0.05.
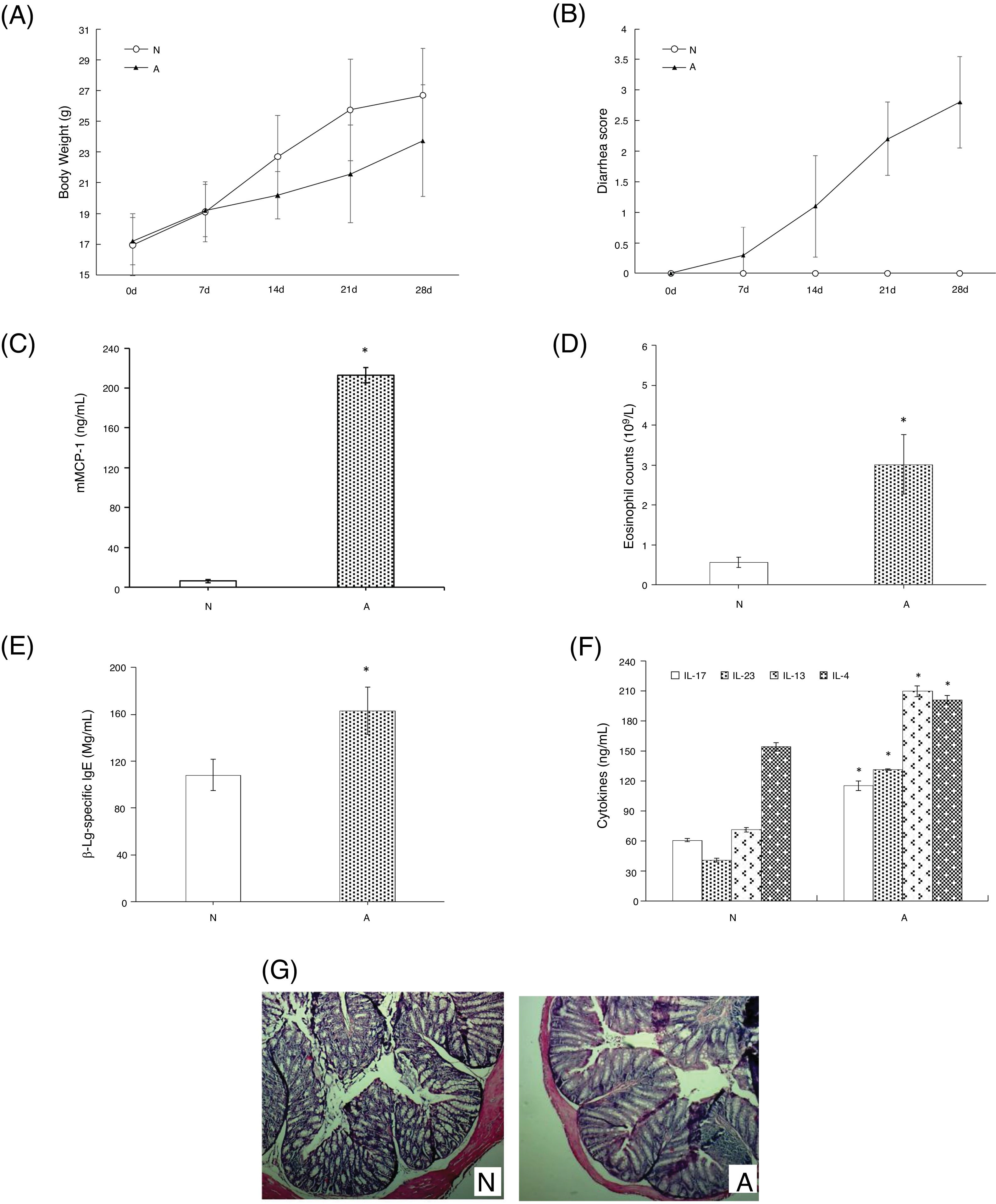
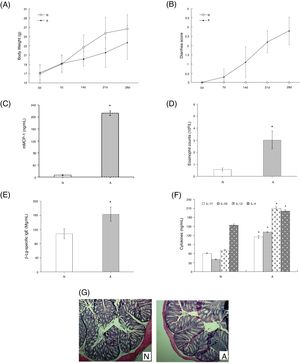
ResultsAssessment of the allergic symptoms of β-Lg-sensitized miceCompared with the control, the allergic mice exhibited a higher diarrhea score and a lower weight growth rate (Fig. 1A,B). In addition, successive β-Lg challenges led to the development of inflammatory response, characterized by a rapid increase in serum mMCP-1 levels and blood eosinophil counts (Fig. 1C,D), as well as enhanced inflammatory cells infiltration and reduced number of crypts in the colon (Fig. 1G). Moreover, high levels of β-Lg specific IgE, Th2-related cytokines (IL-4, IL-13) and Th17-related cytokines (IL-17, IL-6) were observed in the allergic group, which indicated β-Lg promoting Th2 and Th17 dominated inflammation (Fig. 1E,F).
β-Lg-induced food allergy symptoms in 28 d for 1 h after challenge with β-Lg. Body weight (A) and diarrhea score (B) was measured 1 h after every challenge with β-Lg; mMCP-1 levels (C), eosinophil count in blood (D), serum immunoglobulin levels (E), serum Th17 cytokines(IL-17 and IL-23) and Th2 cytokines(IL-4 and IL-13) (F), colon H&E staining (magnification*100) (G) were measured after being sacrificed. Each value is presented as mean ± SD. N: normal group; A: allergic group. (*P < 0.05 vs. normal group).
After removing low-quality sequences, a total of 8,295,663 valid reads were obtained from β-Lg-sensitized and control samples. The length of the RNAs showed that the six libraries shared a similar size distribution, most of which were 20–24 nt long, and the 22-nt small RNAs were the majority size in each library (Fig. S1).
Identification of conserved and novel miRNAsA total of 904 known miRNAs and 189 novel miRNAs were identified in β-Lg and control groups. Among these known miRNAs, 437 of them were found as conserved mature miRNAs, and a total of 963 and 879 known miRNAs were obtained from the allergic group and control, respectively. Venn diagrams results indicated that 749 miRNAs in the overlap of the two groups could be obtained for further investigation (Fig. S2).
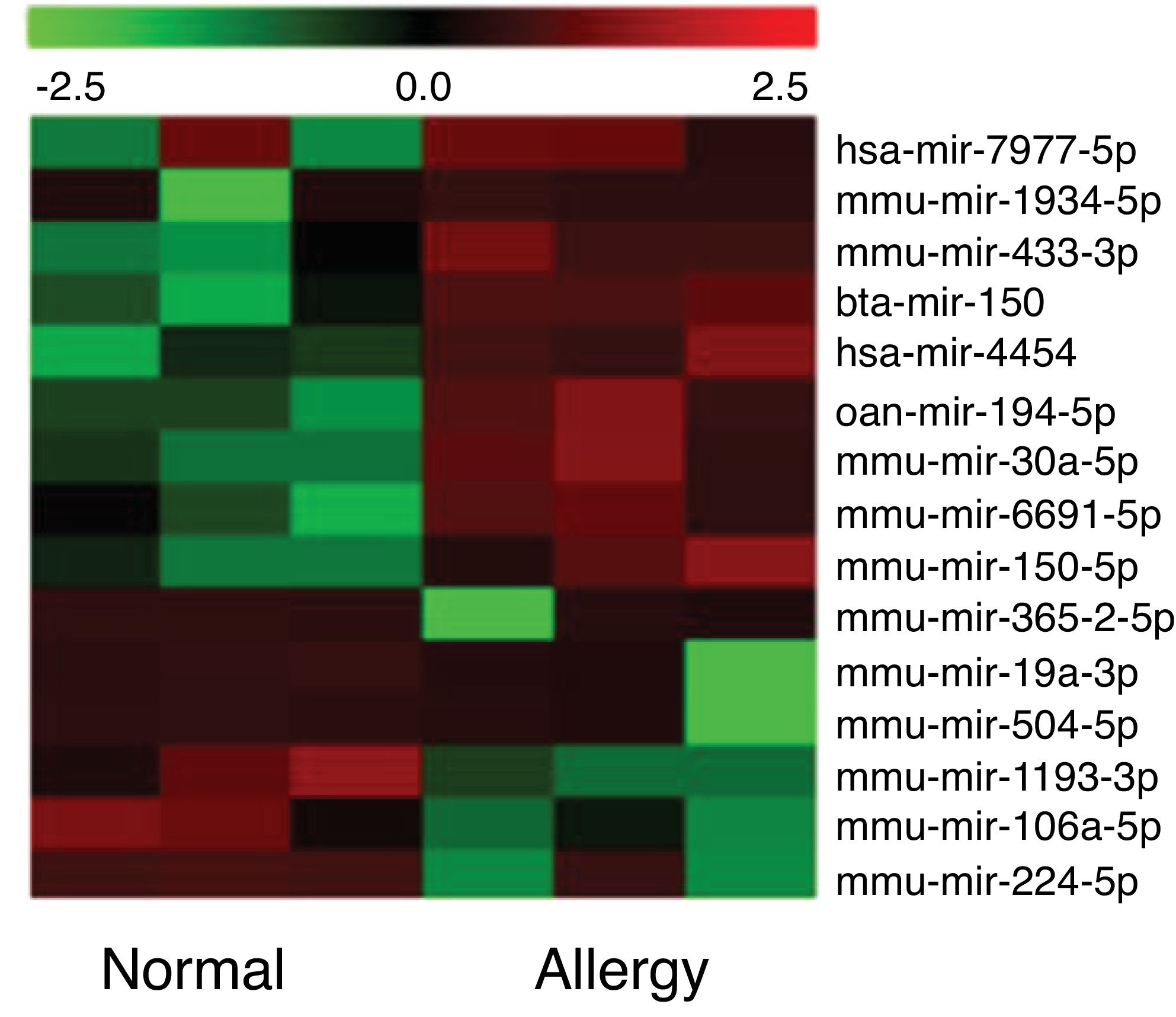
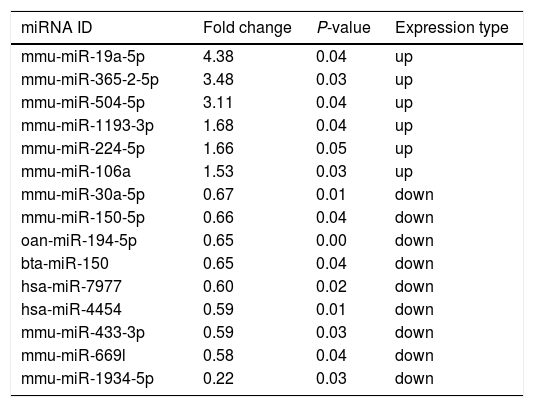
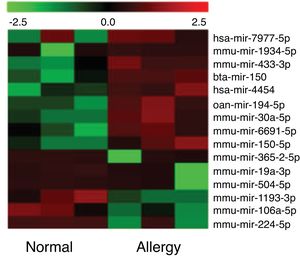
Altered miRNA profiles in colon respond to allergen exposureBy setting the fold change and P-value as threshold, it was found that a total of 16 miRNAs were differentially expressed between the control and β-Lg-treated groups (Fig. 2). Among them, six miRNAs were significantly up-regulated (mmu-miR-19a-3p, mmu-miR-365-2-5p, mmu-miR-504-5p, mmu-miR-1193-3p, mmu-miR-224-5p and mmu-miR-106a) and nine miRNAs were significantly down-regulated (ssc-miR-30a-5p, mmu-miR-150-5p, oan-miR-194-5p, bta-miR-150, hsa-miR-7977, hsa-miR-4454, mmu-miR-669 l, mmu-miR-433-3p and mmu-miR-1934-5p). As shown in Table 2, mmu-miR-19a-5p (fold change = 4.38) and mmu-miR-1934-5p (fold change = 0.22) were the most extremely up-and down-regulated miRNAs, respectively.
Cluster analysis of miRNA differential expression level among the six libraries. Heatmap of differentially expressed miRNAs between the normal group and allergic group. Red and green represent decreased and increased expression, respectively. Normal: normal group; Allergy: allergic group.
Significant differential expression miRNA in two groups.
| miRNA ID | Fold change | P-value | Expression type |
|---|---|---|---|
| mmu-miR-19a-5p | 4.38 | 0.04 | up |
| mmu-miR-365-2-5p | 3.48 | 0.03 | up |
| mmu-miR-504-5p | 3.11 | 0.04 | up |
| mmu-miR-1193-3p | 1.68 | 0.04 | up |
| mmu-miR-224-5p | 1.66 | 0.05 | up |
| mmu-miR-106a | 1.53 | 0.03 | up |
| mmu-miR-30a-5p | 0.67 | 0.01 | down |
| mmu-miR-150-5p | 0.66 | 0.04 | down |
| oan-miR-194-5p | 0.65 | 0.00 | down |
| bta-miR-150 | 0.65 | 0.04 | down |
| hsa-miR-7977 | 0.60 | 0.02 | down |
| hsa-miR-4454 | 0.59 | 0.01 | down |
| mmu-miR-433-3p | 0.59 | 0.03 | down |
| mmu-miR-669l | 0.58 | 0.04 | down |
| mmu-miR-1934-5p | 0.22 | 0.03 | down |
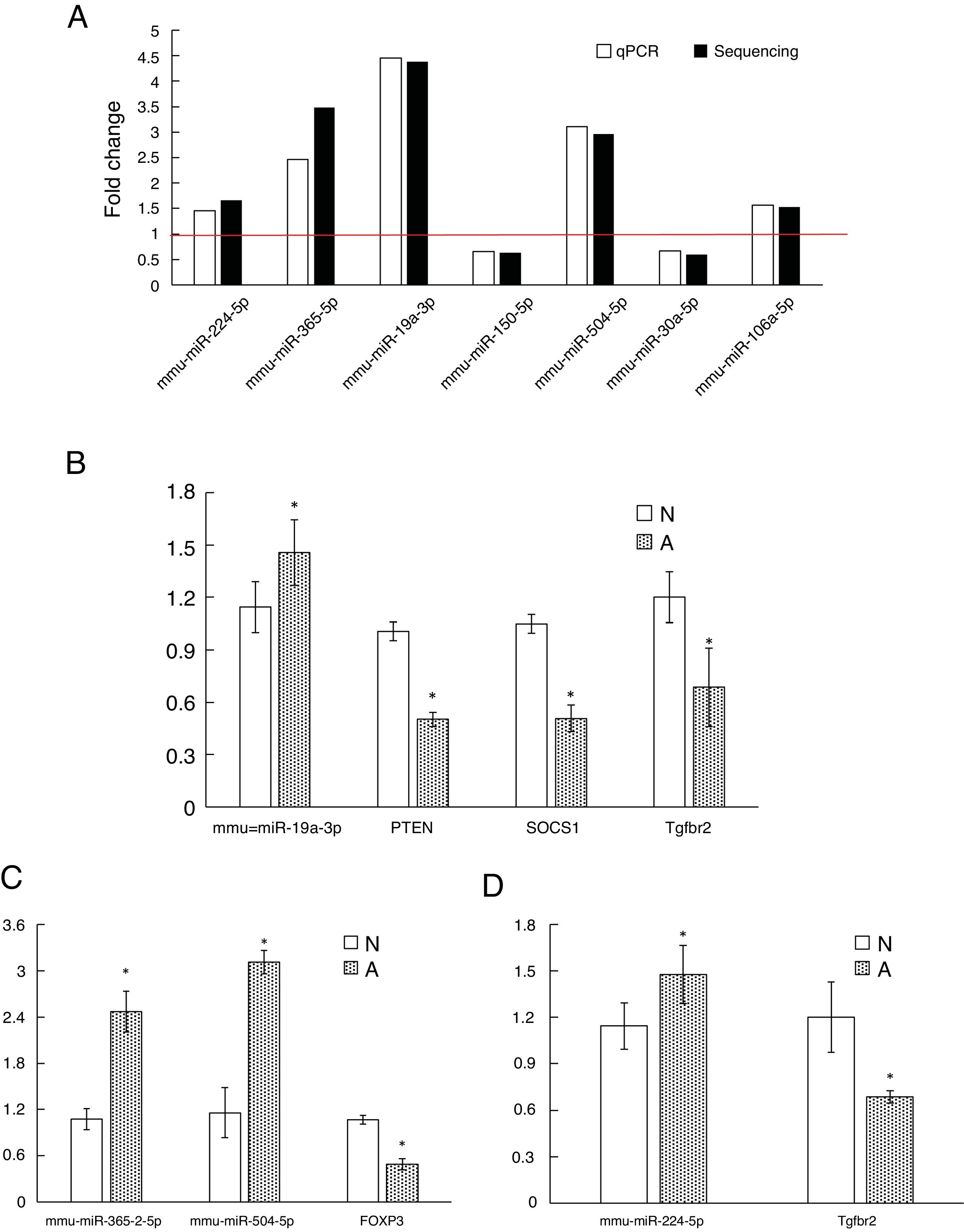
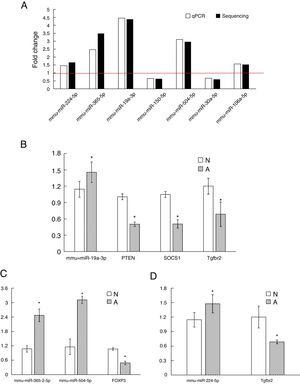
As shown in Fig. 3A, two down-regulated miRNAs (miR-150-5p and miR-30a-5p) and five up-regulated miRNAs (miR-224-5p, miR-365-2-5p, miR-19a-3p, miR-504-5p and miR-106a-5p) were confirmed by qRT-PCR. The verification result was consistent with the sequencing results. The relative fold changes of these miRNAs suggested that the RNA-seq and the abundance results were reliable.
The qRT-PCR validation of seven differential expression miRNAs and eight target genes. (A): The key microRNAs which have a differential expression in colon of β-Lg-sensitized mice compared with control. (B): Target genes of miR-19a-3p. (C): Target genes of miR-365-2-5p and miR-504-5p. (D): Target genes of miR-224-5p. N: normal group; A: allergic group. (*P < 0.05 vs. normal group).
In order to research the biological function of these specific miRNA taking part in β-Lg allergy, target genes were predicted by TargetScan and miRanda, the intersection of the two software packages was taken as the final target gene. A total of 70,589 predicted targets were obtained, which were clustered into 1941 significantly enriched GO terms. Fig. S3 A showed the 50 most significantly enriched GO terms. In the biological process, most target genes were correlated with transcription, DNA-templated and regulation of transcription. Among the GO terms for the cellular component, cytoplasm, membrane and nucleus were enriched by massive miRNAs. Protein binding and metal ion binding were the most extremely enriched terms in molecular function.
The KEGG analysis showed that the significantly enriched pathways were pathways in cancer (Fig. S3 B). KEGG cancer pathways (ko05200) contained members of different signaling pathway members within a single pathway, e.g. the NF-κB, JAK/STAT, TGF-β, PI3K/AKT and cAMP signaling.19
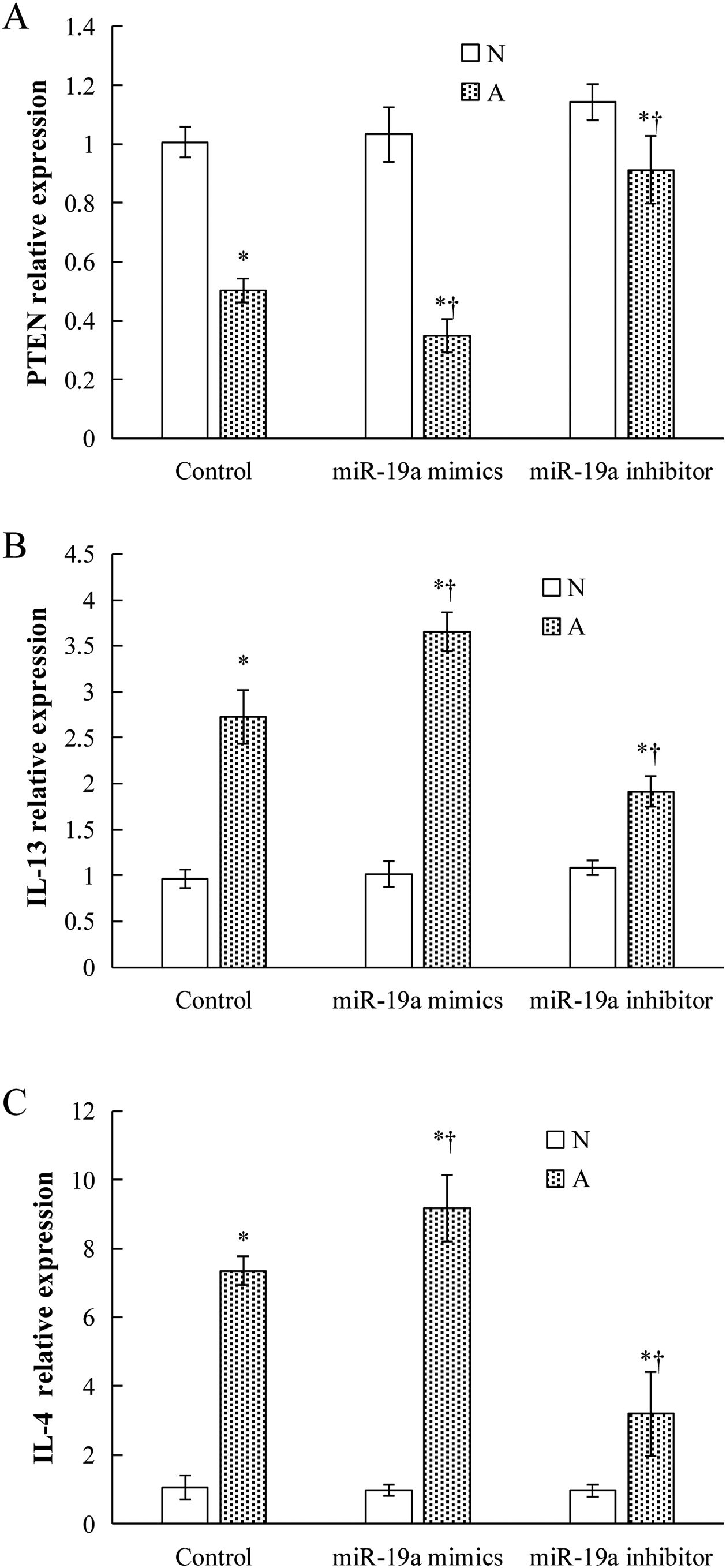
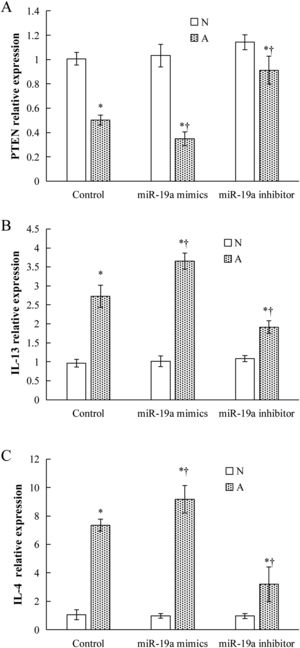
Validation of key candidate miRNAs and target genesAs shown in Fig. 3B–D, four allergic inflammatory related genes (PTEN, SOCS1, Tgfbr2, FOXP3) were inhibited in the β-Lg-allergic group, and presented the opposite trend with their candidate miRNAs (miR-19a-3p, miR-504-5p, miR-224-5p and miR-365-2-5p). These negative relationships of six miRNAs-gene pairs (miR-19a/PTEN, miR-19a/SOCS1, miR-19a/Tgfbr2, miR365-2/FOXP3, miR-504/FOXP3, miR-224/Tgfbr2) was concordant with the target gene prediction results. To further explore the interaction between the predicted binding sites for miR-19a within its target gene, RAW264.7 cells were transfected with miR-19a mimics and inhibitor. qRT-PCR analysis confirmed that the cells transfected with miR-19a mimiced significantly attenuated PTEN expression, while miR-19a inhibitor treatment leads to opposite results (Fig. 4 by miR-19a mimic transfection and reduced by miR-19a inhibitor transfection (Fig. 4B,C). These results indicated the important role of miR-19a in promoting Th2-mediated inflammation.
DiscussionMiRNAs have recently been recognized as powerful regulators of host gene expression, and played a determinant role in the process of innate and adaptive immunity. As potential biomarkers, most evidence showed the altered miRNA profiles present in inflammatory and autoimmune diseases. In the present investigation, a total of 15 differentially expressed miRNAs were detected in the colon of β-Lg allergic mice and normal mice, including six up-regulated miRNAs such as miR-19a-3p, miR-365-2-5p, miR-504-5p, miR-224-5p, miR-106a and 9 down-regulated miRNA such as miR-30a-5p, miR-150-5p, miR-433-3p. Among them, miR-106a and miR-150 were previously identified to participate in hypersensitivity. For example, the increase of miR-106a could inhibit IL-10 secretion in mice with allergic airway inflammation, while the knockdown of miR-106a helped to alleviate numerous asthma reactions.20 Additionally, miR-150, which was decreased in β-Lg-sensitized mice, displayed an identical trend in an asthma rat model and targeted BCYRN1 which is a crucial modifier of airway smooth muscle cells proliferating and migrating.21
Our previous study has confirmed that β-Lg allergy was characterized by predominant Th17/Treg imbalance, along with abnormal Th2 cell response.22 Two miRNAs in the current study, the up miR-19a and down miR-30a, were shown to have a potential role in allergen-specific T cells response. miR-17∼92 cluster including miR-19a was identified as a pleiotropic regulator of Th2 cell response.23 Simpson et al. provided evidence that elevated miR-19a in human airway-infiltrating T cells effectively initiated Th2 cytokines production (IL-4, IL-13 and IL-5) by negatively regulating Th2 limiting factors, such as PTEN, SOCS1 and A20,24 this targeted regulatory relationship was confirmed in the transfection experiment of this study. On the contrary, miR-30a, which was down-regulated by 1.5 times, has an inverse correlation with Th17 phenotype and Th17-polarising cytokine (IL-17a) expression. Zhao et al. indicated that miR-30a targeted with IRF4, a key transcriptional factor of Th17 differentiation, which led to attenuate Th17-induced autoimmune disease.25 Besides, an increase in miR-30a could weaken IL-17-related inflammation by targeting adaptor of IL-17 signaling-Act1.26
Our prophase research also proved that the IL-6/STAT3 pathway was indispensable to maintain Th17/Treg imbalance in β-Lg sensibilization.27 Next, the observation revealed that the target genes of altered miRNAs were involved in various pathways that stimulated CD4+ cell differentiation, such as JAK/STAT and NF-κB signaling. For instance, the role of miR-106a, a significant down-regulated miRNA in the present study, has been identified to negatively adjust STAT3. Zhang et al. showed that miR-106a reduced STAT3 and phospho-STAT3 expression via transfection of human neuroblastoma SH-SY5Y cells.20 Similarly, He et al. demonstrated that miR-106a targeted STAT3 was beneficial to renewal of spermatogonial stem cells in mice.28 Accordingly, we speculated that the depletion of miR-106a might potentially promote the signal transduction of JAK/STAT3 during the allergic process. As another pathway required by Th2 and Th17 proliferation, NF-κB signaling could be activated by miR-19a and miR-224. Jun Hong et al. found that high expressed miR-19a led to elevated NF-κB transcription in oxidative stress-initiated cell.29 This results were also supported by Renli Qi et al.’s finding that over-expression of miR-224 served to prevent the adipocyte apoptosis by promoting NF-κB signaling.30
Conversely, patients with food allergy generally had allergens tolerance failure followed by diminishing of Treg cell. Notably, our predicted target genes were also enriched into the pathways and genes that were closely related to Treg deficiency. Firstly, TGF-β served as a representative cytokine of Treg cell, has an important anti-allergic mediatory characteristic, and decreased TGF-β signaling pathway activity would result in Treg function defects in anaphylactic reaction.31 TGF-beta effect was clearly dictated by composition of TGF-beta receptor type II (Tgfbr2) and type I (Tgfbr1).32 In this study, both miR-19a and miR-224 targeted Tgfbr2 and their co-regulation results have been confirmed by qPCR. Similarly, Haj-Salem et al. suggested that active expressing miR-19a targeted Tgfbr2 and inhibited TGF-β signaling pathway in human bronchial epithelial cells from asthma patients.33 Second, the importance of the PI3K/AKT signaling pathway in natural Treg development has been documented.34 Okkenhaug et al. observed that the proportion of Tregs were increased in the PI3K-kinase-inactive mice and speculated that PI3K had the ability to inhibit Treg response.35 The MiR-17-92 cluster was documented as inducing PI3K/AKT arise,36 which suggested that miR-19a accumulation in the colon during β-Lg challenge might exert its Treg-suppressing function. Thirdly, FOXP3 encoded the transcription factor that drove the development and function of Tregs, and the decrease in FOXP3 led to Th17 cell polarization.37 In this research, FOXP3 was observed to regulate by multiple miRNAs (such as miR-365-2, miR-504) and these co-regulation results of miRNA and mRNA were further confirmed by qPCR. MiR-504 has been shown to be a biological target affecting tumor cell chemoresistance,38 while its potential effect on impairing Treg function via suppressing FOXP3 transcription has still not been thoroughly investigated. Accordingly, more studies are required to explore these corresponding regulatory mechanisms.
ConclusionTaken together, β-Lg challenge led to promoting Th2 and Th17 dominant allergic inflammatory, together with specific miRNA profiles. Function annotation of the predicted target genes of altered miRNAs revealed a broad range of pathways and processes related to immune dysregulation and allergic inflammation. These results would provide valuable information for further investigation on the miRNA-mediated regulation in CMA.
Conflict of interestThe authors declare that they have no conflict of interest.
We gratefully acknowledge the financial support of the National Key Research and Development Program of China (grant number 2016YFD0400605 and 2017YFD0400304).