Allergy and autoimmunity are important immunological entities underlying chronic diseases in children. In some cases both entities develop simultaneously in the same patient. FOXP3 gene codes for a transcription factor involved in regulation of the immune system.
Considering that regulatory T cells are involved in controlling immunological disease development, and the relevant role of FOXP3 in this kind of T cells, the objective of this study was to analyse the FOXP3 gene in the most prevalent autoimmune diseases and/or allergies in childhood in a European population.
MethodsA total of 255 Caucasian individuals, 95 controls and 160 patients diagnosed with allergic, autoimmune or both diseases were included in this study. The molecular analysis of FOXP3 was performed by DNA sequencing following the recommendations for quality of the European Molecular Genetics Quality Network. Genomic DNA was extracted from peripheral blood of all participants and was amplified using the polymerase chain reaction. After the visualisation of the amplified fragments by agarose gel-electrophoresis, they were sequenced.
ResultsThirteen different polymorphisms in FOXP3 gene were found, seven of which had not been previously described. The mutated allele of SNP 7340C>T was observed more frequently in the group of male children suffering from both allergic and autoimmune diseases simultaneously (p=0.004, OR=16.2 [1.34–195.15]).
ConclusionsIn this study we identified for first time genetic variants of FOXP3 that are significantly more frequent in children who share allergic and autoimmune diseases. These variants mainly affect regulatory sequences that could alter the expression levels of FOXP3 modifying its function including its role in Treg cells.
Chronic diseases affecting children produce a significant impairment in their quality of life, an alteration in familial and social relationships, and an increase in morbidity and mortality. Many of them are of immune origin, such are allergic and autoimmune diseases. Allergies are due to an immunological response to a usually innocuous antigen (allergen) resulting in the proliferation of T-helper type 2 (Th2) lymphocytes. Autoimmune diseases are produced by an immune response with proliferation of Th1 lymphocytes, which react against own antigens recognised as foreign (autoantigens). Genetic and environmental factors participate in their pathogenic mechanisms. Asthma, rhinoconjunctivitis (RC) and atopic dermatitis (AD) are the most prevalent allergic diseases, and type 1 diabetes mellitus (DM1), coeliac disease (CD) and autoimmune thyroiditis (AT) stand out among autoimmune diseases.
The IPEX syndrome (immunodysregulation, polyendocrinopathy, enteropathy, X-linked), also called XLAAD (X-linked autoimmunity-allergic disregulation), encompasses both immune pathologies. Forkhead Box P3 (FOXP3) was identified as a gene involved in this syndrome.1 However, in some patients with IPEX symptoms no mutations have been identified in the FOXP3 coding region being diagnosed as IPEX-like syndrome.2,3 Mutations have been described in the regulatory regions in some of them.2
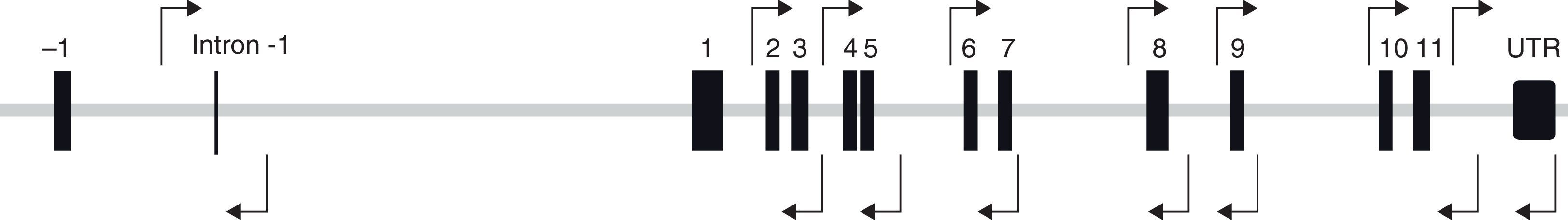
FOXP3 gene, also called JM2, is located in Xp11.23 and has eleven coding and three non-coding exons (Fig. 1). It is expressed mainly in a subgroup of CD4+ cells, called T regulatory cells (Treg).4 The FOXP3 protein is the most important marker of this cell subtype. Its function is not completely established, but it is involved in Treg cell activation.5,6
Treg cells represent 5–10% of CD4+ T cells.7 They also express the alpha chain of the IL-2 receptor, CD25. In addition, cytotoxic T-lymphocyte antigen 4 (CTLA-4) and glucocorticoid induced tumour necrosis factor receptor family related gene (GITR) are also important for Treg function.4,5 These Treg cells regulate the innate and adaptive immunity, thus contributing to the maintenance of immune tolerance.6,7 When their function is affected, autoimmune and allergic diseases can appear. Moreover, Treg cells regulate the immune system involved in host defence against infection and tumour proliferation.7,8 The mechanisms involved in these immunosuppressive effects are not well elucidated; some studies suggest they are cell contact-dependent,9,10 while others report them to be cell contact-independent.10 FOXP3 is thought to interact to DNA assisted by FOXP3 cofactors orchestrating a transcription network by multimerisation. Thus FOXP3 functions may change depending on the partners forming the so-called FOXP3 interactome. FOXP3 is considered a multifaceted transcription factor that regulates immune homeostasis.11 It stimulates the production of inhibitory cytokines, like interleukin-10 (IL-10) and tumour growth factor-β (TGF-β), suppressing IL-2 and IL-4. When this regulation is not present, autoimmunity and allergy can emerge.5,7
Considering that Treg cells are involved in controlling the development of autoimmune and allergic diseases, and the relevant role of FOXP3 in these cells, our hypothesis was that FOXP3 could be involved in the development of immune diseases and the objective of this study was to analyse the FOXP3 gene for polymorphisms associated with the most prevalent autoimmune and/or allergic diseases in children.
Materials and methodsParticipantsThis is an observational, case-control study in which 255 non-related Caucasian individuals were included: 95 controls and 160 patients that were recruited at the Allergy and Paediatric Departments of the University Hospital of Salamanca. The study was performed following the recommendations of the Hospital's Ethics Committee. An informed written consent was obtained from the children's parents or caregivers.
All patients were under 18 years old, and were affected by one or more autoimmune diseases (DM1, AT and/or CD) and/or atopy. Diagnosis was according to World Health Organization and American Diabetes Association criteria for DM1;12 thyrotropin up to 10μU/mL plus thyroid autoantibodies for AT;13 the European Society for Paediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN) criteria for CD;14 atopy was defined by a positive skin prick test to at least one of a battery of common aeroallergens previously described;15 in addition, patients had rhinoconjunctivitis, and/or asthma and/or atopic dermatitis diagnosed by an allergologist.
Controls were selected from adults, confirming the absence of atopy or autoimmune disease during childhood and adulthood.16 The following inclusion criteria were required: (i) absence of autoimmune diseases; (ii) absence of clinical symptoms compatible with bronchial asthma or other respiratory diseases; (iii) absence of rhinoconjunctivitis or atopic dermatitis; (iv) absence of other allergic diseases; and (v) negative skin prick tests to the same battery of common aeroallergens employed with patients.
Molecular analysis of FOXP3 geneA molecular analysis of FOXP3 was carried out in all subjects included in the study. All laboratory procedures were developed following the recommendations of quality of European Molecular Genetics Quality Network.17 Genomic DNA was extracted from peripheral blood with MagNA Pure Compact System (Roche, Barcelona, Spain). FOXP3 regions were amplified using the polymerase chain reaction (PCR) (Fig. 1).
The pair of primers ATCTCCATGAGCCTCAGTTTC and CGTTTTTTGGAGGGTGGA were used to amplify exons 2 and 3, AGTCAAAATGTCAGCACCTG and ATGGGGGTAAATAACAGCAC for exons 4 and 5, AGAGGGAGACTGAGGTAGAGAG and TGAATGTGAGGTTAGGTTCC for exons 6 and 7, AGGCTGAGGCAGAAGACTTG and CCAAGGGAGTCAGGGCTA for exon 8, CTGGACACGGGTGTTGACGG and GGCACTCAGAGGGAGACAGG for exon 9, ATGTCAGATGGCTCGGGGTAG and TCAGGGCTGTGCTTGTGTG for exons 10 and 11, GCCAAACAGAGCCTTCACAAC and GTGATGGGGGCTGGTGAG for 3′UTR region and TCGATTCTGCAGGCTTAGCA and GGAAATGGAGGTATGGAGAGGT to amplify intron −1. The PCR was carried out in an MWG-BIOTECH thermocycler (Ebersberg, Germany). The amplification programme included one cycle of 95°C for 5min; 35 cycles at 94°C for 30s, annealing temperature (60–67.5°C) of each reaction for 1min and 72°C for 1min; and a extension period at 72°C for 5min. The annealing temperature was established depending on annealing temperature of the pair of primers used: 60°C for exons 4, 5, 6 and 7; 61°C for exons 2 and 3; 63°C for exon 8; 64°C for intron −1; 65°C for exon 9; and 67.5°C for exons 10 and 11 and 3′UTR region. Following PCR, the amplified fragments were visualised by horizontal agarose gel-electrophoresis using Redsafe (iNtRON Biotechnology Inc., Korea). The PCR products were purified using ExoSAP-IT (Affymetrix, Barcelona, Spain) or IllustraExoSTAR 1-step or 2-steps (VWR International Eurolab, Barcelona, Spain) and sequenced in a 3100 Genetic Analyzer Sequencer using the same PCR primers (Applied Biosystems, CA, USA).
Bioinformatic analysisNG_007392.1, the reference sequence of FOXP3, was identified by OMIM platform (http://www.ncbi.nlm.nih.gov/omim) and polymorphisms were sought in an on-line platform NCBI (http://www.ncbi.nlm.nih.gov/snp). Primers were designed with VectorNTI 10.3.0© 2006 Invitrogen Corporation (Carlsbad, California, USA) and they were optimised with the on-line programmes Beacon Designer (http://www.premierbiosoft.com/qpcr/index.html) and Netprimer (http://premierbiosoft.com/netprimer/index.html). Chromas 2.3 (Technelysium. Pty. Ltd. 1998–2004, South Brisbane, Australia) was used to visualise chromatograms of each sequenced fragment; and sequence alignments were then performed using Vector NTI to compare with the reference sequence of FOXP3, NG_007392.1. ExPASy software platform (http://web.expasy.org/translate) was used for protein predictions.
Statistical analysisA descriptive analysis was performed using percentages for qualitative variables, and measures of central tendency (mean) and dispersion measures (standard deviation) for quantitative variables. The allele, genotype and haplotype frequencies were compared in patients and controls. As the FOXP3 gene is located on chromosome X males and females were separately analysed. These frequencies were compared between groups using χ2, Pearson and Fisher's exact test. Odds ratios (95% Confidence Interval) were calculated. The Hardy–Weinberg equilibrium was tested by a χ2 test (Pearson's P-value). To avoid type I error, the statistical power and the false positive report probability (FPRP) were calculated, and a more astringent p value of 0.01 was considered statistically significant instead of 0.05. Bonferroni was applied when proceeded. To analyse the data the software package SPSS version 18.0 (Chicago, Illinois, USA) and SHEsis software platform (http://analysis.bio-x.cn/myAnalysis.php) were employed.
ResultsAs stated, 255 unrelated Caucasian individuals were included, 95 controls and 160 patients, 43.2% of controls (mean age of 53.7±16.9) and 55.6% of patients (mean age 8.6±3.9) were males; 76 (47.5%) patients were atopic, 72 (45%) had an autoimmune disorder and 12 (7.5%) had both atopy and autoimmunity.
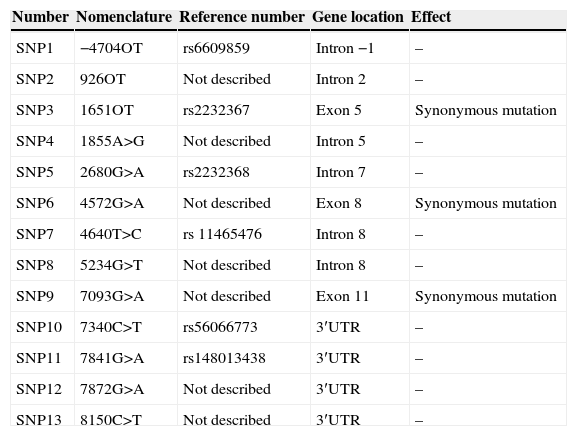
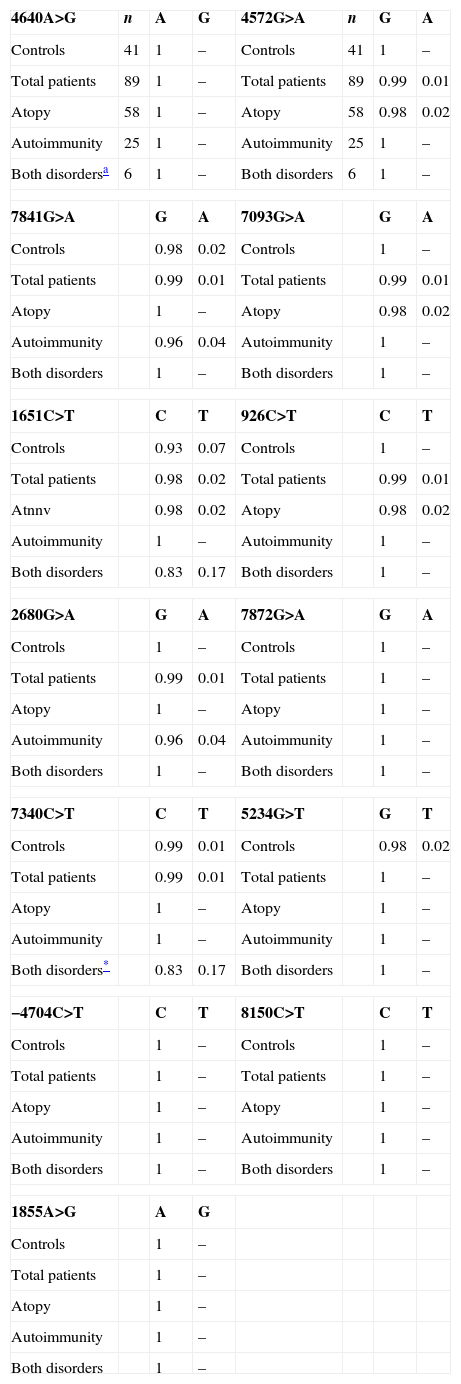
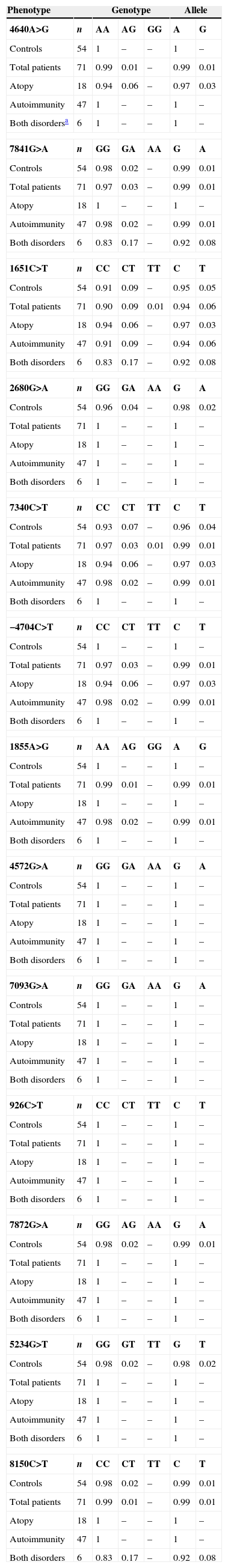
In the FOXP3 gene, 13 different polymorphisms were found (Table 1), seven of which had not been previously described. Gene frequencies of these SNPs, separated by gender, were calculated in both control and case groups (Tables 2 and 3). Hardy–Weinberg equilibrium was accomplished in all groups. Statistically significant differences were observed, especially in the male children with both autoimmune and atopic diseases. There was an increase in the presence of the mutated allele in the position 7340C>T, specifically in those patients that suffered from both atopic and autoimmune diseases (p=0.004, OR=16.2 [1.34–195.15]), statistical power of 75%, with a false positive report probability (FPRP) of 7%. Meanwhile, the mutated allele in position 1651C>T was more frequent in controls (p=0.01, OR=0.16 [0.03–0.79]) than in patients who had only one disease, i.e. atopy or autoimmunity (statistical power of 77.7% and FPRP of 6.2%) (Table 2). No statistically significant differences were observed in females (Table 3).
Polymorphisms observed in the molecular analysis of the FOXP3 gene.
| Number | Nomenclature | Reference number | Gene location | Effect |
|---|---|---|---|---|
| SNP1 | −4704OT | rs6609859 | Intron −1 | – |
| SNP2 | 926OT | Not described | Intron 2 | – |
| SNP3 | 1651OT | rs2232367 | Exon 5 | Synonymous mutation |
| SNP4 | 1855A>G | Not described | Intron 5 | – |
| SNP5 | 2680G>A | rs2232368 | Intron 7 | – |
| SNP6 | 4572G>A | Not described | Exon 8 | Synonymous mutation |
| SNP7 | 4640T>C | rs 11465476 | Intron 8 | – |
| SNP8 | 5234G>T | Not described | Intron 8 | – |
| SNP9 | 7093G>A | Not described | Exon 11 | Synonymous mutation |
| SNP10 | 7340C>T | rs56066773 | 3′UTR | – |
| SNP11 | 7841G>A | rs148013438 | 3′UTR | – |
| SNP12 | 7872G>A | Not described | 3′UTR | – |
| SNP13 | 8150C>T | Not described | 3′UTR | – |
Distribution of allelic frequencies in males.
| 4640A>G | n | A | G | 4572G>A | n | G | A |
| Controls | 41 | 1 | – | Controls | 41 | 1 | – |
| Total patients | 89 | 1 | – | Total patients | 89 | 0.99 | 0.01 |
| Atopy | 58 | 1 | – | Atopy | 58 | 0.98 | 0.02 |
| Autoimmunity | 25 | 1 | – | Autoimmunity | 25 | 1 | – |
| Both disordersa | 6 | 1 | – | Both disorders | 6 | 1 | – |
| 7841G>A | G | A | 7093G>A | G | A | ||
| Controls | 0.98 | 0.02 | Controls | 1 | – | ||
| Total patients | 0.99 | 0.01 | Total patients | 0.99 | 0.01 | ||
| Atopy | 1 | – | Atopy | 0.98 | 0.02 | ||
| Autoimmunity | 0.96 | 0.04 | Autoimmunity | 1 | – | ||
| Both disorders | 1 | – | Both disorders | 1 | – | ||
| 1651C>T | C | T | 926C>T | C | T | ||
| Controls | 0.93 | 0.07 | Controls | 1 | – | ||
| Total patients | 0.98 | 0.02 | Total patients | 0.99 | 0.01 | ||
| Atnnv | 0.98 | 0.02 | Atopy | 0.98 | 0.02 | ||
| Autoimmunity | 1 | – | Autoimmunity | 1 | – | ||
| Both disorders | 0.83 | 0.17 | Both disorders | 1 | – | ||
| 2680G>A | G | A | 7872G>A | G | A | ||
| Controls | 1 | – | Controls | 1 | – | ||
| Total patients | 0.99 | 0.01 | Total patients | 1 | – | ||
| Atopy | 1 | – | Atopy | 1 | – | ||
| Autoimmunity | 0.96 | 0.04 | Autoimmunity | 1 | – | ||
| Both disorders | 1 | – | Both disorders | 1 | – | ||
| 7340C>T | C | T | 5234G>T | G | T | ||
| Controls | 0.99 | 0.01 | Controls | 0.98 | 0.02 | ||
| Total patients | 0.99 | 0.01 | Total patients | 1 | – | ||
| Atopy | 1 | – | Atopy | 1 | – | ||
| Autoimmunity | 1 | – | Autoimmunity | 1 | – | ||
| Both disorders* | 0.83 | 0.17 | Both disorders | 1 | – | ||
| −4704C>T | C | T | 8150C>T | C | T | ||
| Controls | 1 | – | Controls | 1 | – | ||
| Total patients | 1 | – | Total patients | 1 | – | ||
| Atopy | 1 | – | Atopy | 1 | – | ||
| Autoimmunity | 1 | – | Autoimmunity | 1 | – | ||
| Both disorders | 1 | – | Both disorders | 1 | – | ||
| 1855A>G | A | G | |||||
| Controls | 1 | – | |||||
| Total patients | 1 | – | |||||
| Atopy | 1 | – | |||||
| Autoimmunity | 1 | – | |||||
| Both disorders | 1 | – | |||||
(–) Not detected.
Distribution of genotypic and allelic frequencies in females.
| Phenotype | Genotype | Allele | ||||
|---|---|---|---|---|---|---|
| 4640A>G | n | AA | AG | GG | A | G |
| Controls | 54 | 1 | – | – | 1 | – |
| Total patients | 71 | 0.99 | 0.01 | – | 0.99 | 0.01 |
| Atopy | 18 | 0.94 | 0.06 | – | 0.97 | 0.03 |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disordersa | 6 | 1 | – | – | 1 | – |
| 7841G>A | n | GG | GA | AA | G | A |
| Controls | 54 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Total patients | 71 | 0.97 | 0.03 | – | 0.99 | 0.01 |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Both disorders | 6 | 0.83 | 0.17 | – | 0.92 | 0.08 |
| 1651C>T | n | CC | CT | TT | C | T |
| Controls | 54 | 0.91 | 0.09 | – | 0.95 | 0.05 |
| Total patients | 71 | 0.90 | 0.09 | 0.01 | 0.94 | 0.06 |
| Atopy | 18 | 0.94 | 0.06 | – | 0.97 | 0.03 |
| Autoimmunity | 47 | 0.91 | 0.09 | – | 0.94 | 0.06 |
| Both disorders | 6 | 0.83 | 0.17 | – | 0.92 | 0.08 |
| 2680G>A | n | GG | GA | AA | G | A |
| Controls | 54 | 0.96 | 0.04 | – | 0.98 | 0.02 |
| Total patients | 71 | 1 | – | – | 1 | – |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 7340C>T | n | CC | CT | TT | C | T |
| Controls | 54 | 0.93 | 0.07 | – | 0.96 | 0.04 |
| Total patients | 71 | 0.97 | 0.03 | 0.01 | 0.99 | 0.01 |
| Atopy | 18 | 0.94 | 0.06 | – | 0.97 | 0.03 |
| Autoimmunity | 47 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Both disorders | 6 | 1 | – | – | 1 | – |
| −4704C>T | n | CC | CT | TT | C | T |
| Controls | 54 | 1 | – | – | 1 | – |
| Total patients | 71 | 0.97 | 0.03 | – | 0.99 | 0.01 |
| Atopy | 18 | 0.94 | 0.06 | – | 0.97 | 0.03 |
| Autoimmunity | 47 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 1855A>G | n | AA | AG | GG | A | G |
| Controls | 54 | 1 | – | – | 1 | – |
| Total patients | 71 | 0.99 | 0.01 | – | 0.99 | 0.01 |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 4572G>A | n | GG | GA | AA | G | A |
| Controls | 54 | 1 | – | – | 1 | – |
| Total patients | 71 | 1 | – | – | 1 | – |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 7093G>A | n | GG | GA | AA | G | A |
| Controls | 54 | 1 | – | – | 1 | – |
| Total patients | 71 | 1 | – | – | 1 | – |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 926C>T | n | CC | CT | TT | C | T |
| Controls | 54 | 1 | – | – | 1 | – |
| Total patients | 71 | 1 | – | – | 1 | – |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 7872G>A | n | GG | AG | AA | G | A |
| Controls | 54 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Total patients | 71 | 1 | – | – | 1 | – |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 5234G>T | n | GG | GT | TT | G | T |
| Controls | 54 | 0.98 | 0.02 | – | 0.98 | 0.02 |
| Total patients | 71 | 1 | – | – | 1 | – |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 1 | – | – | 1 | – |
| 8150C>T | n | CC | CT | TT | C | T |
| Controls | 54 | 0.98 | 0.02 | – | 0.99 | 0.01 |
| Total patients | 71 | 0.99 | 0.01 | – | 0.99 | 0.01 |
| Atopy | 18 | 1 | – | – | 1 | – |
| Autoimmunity | 47 | 1 | – | – | 1 | – |
| Both disorders | 6 | 0.83 | 0.17 | – | 0.92 | 0.08 |
(–) Not detected.
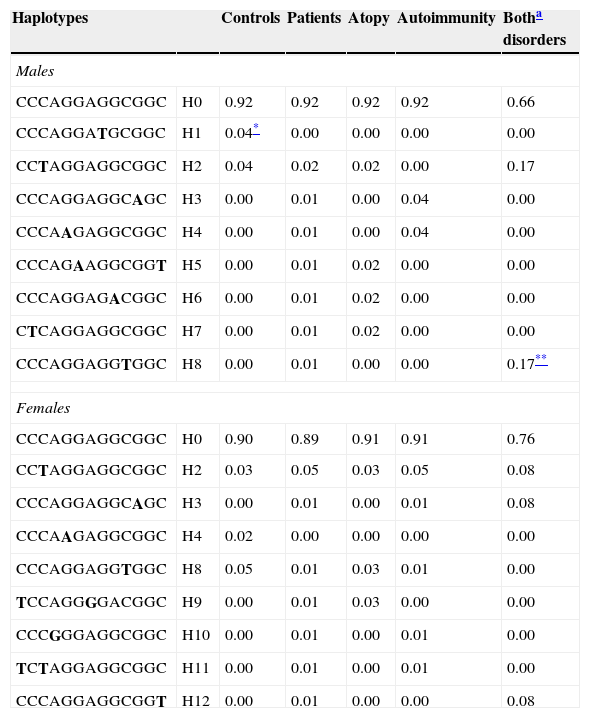
Haplotype combinations and their frequencies were also determined (Table 4). In males, haplotype H8 was only described in patients with both diseases (p=0.006) in contrast to haplotype H1 that was only identified in controls (p=0.005).
Haplotype frequencies. Data represent the percentage of each haplotype in each phenotypic group. Haplotypes with a frequency >1% among either controls or patients are included.
| Haplotypes | Controls | Patients | Atopy | Autoimmunity | Botha disorders | |
|---|---|---|---|---|---|---|
| Males | ||||||
| CCCAGGAGGCGGC | H0 | 0.92 | 0.92 | 0.92 | 0.92 | 0.66 |
| CCCAGGATGCGGC | H1 | 0.04* | 0.00 | 0.00 | 0.00 | 0.00 |
| CCTAGGAGGCGGC | H2 | 0.04 | 0.02 | 0.02 | 0.00 | 0.17 |
| CCCAGGAGGCAGC | H3 | 0.00 | 0.01 | 0.00 | 0.04 | 0.00 |
| CCCAAGAGGCGGC | H4 | 0.00 | 0.01 | 0.00 | 0.04 | 0.00 |
| CCCAGAAGGCGGT | H5 | 0.00 | 0.01 | 0.02 | 0.00 | 0.00 |
| CCCAGGAGACGGC | H6 | 0.00 | 0.01 | 0.02 | 0.00 | 0.00 |
| CTCAGGAGGCGGC | H7 | 0.00 | 0.01 | 0.02 | 0.00 | 0.00 |
| CCCAGGAGGTGGC | H8 | 0.00 | 0.01 | 0.00 | 0.00 | 0.17** |
| Females | ||||||
| CCCAGGAGGCGGC | H0 | 0.90 | 0.89 | 0.91 | 0.91 | 0.76 |
| CCTAGGAGGCGGC | H2 | 0.03 | 0.05 | 0.03 | 0.05 | 0.08 |
| CCCAGGAGGCAGC | H3 | 0.00 | 0.01 | 0.00 | 0.01 | 0.08 |
| CCCAAGAGGCGGC | H4 | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 |
| CCCAGGAGGTGGC | H8 | 0.05 | 0.01 | 0.03 | 0.01 | 0.00 |
| TCCAGGGGACGGC | H9 | 0.00 | 0.01 | 0.03 | 0.00 | 0.00 |
| CCCGGGAGGCGGC | H10 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 |
| TCTAGGAGGCGGC | H11 | 0.00 | 0.01 | 0.00 | 0.01 | 0.00 |
| CCCAGGAGGCGGT | H12 | 0.00 | 0.01 | 0.00 | 0.00 | 0.08 |
The order of the SNPs is −4704C>T, 926C>T, 1651C>T, 1855A>G, 2680G>A, 4572G>A, 4640A>G, 5234G>T, 7093G>A, 7340C>T, 7841G>A, 7872G>A and 8150C>T (this is the order in the gene sequence). Bold letters indicate the mutated allele.
We studied the FOXP3 gene and evaluated allele, genotype and haplotype frequencies in a paediatric population of atopic patients and patients with autoimmune diseases. We identified 13 SNPs, including seven previously undescribed. We found different associations according to gender. These differences between genders have been previously described in other genetic association studies for SNPs in genes located on chromosome X.18
The most relevant difference was detected in the group of participants that simultaneously suffered atopic and autoimmune diseases. The mutated allele of 7340C>T was significantly associated with the simultaneous presence of autoimmunity and atopy in male children, but not those that only suffered from one disease. This polymorphism is located in the 3′ untranslated region, which is described as a regulatory region, since it could modify mRNA stability and translation efficiency.19 In addition, this polymorphism could modify cellular localisation signals affecting protein location, and consequently, FOXP3 function.
We also detected a significant increase in the frequency of the mutated allele of 1651C>T in the male control group. This SNP is located in the coding region (Exon 5) and it is a synonymous one, with no change in protein sequence. Traditionally these changes were considered silent mutations. Nowadays it is thought that it could be involved in transcriptional regulation.20 Polymorphisms may reside in exonic splicing enhancers, near the exonic extreme that defines the point where the splicing ‘machinery’ has to work.21 Another described mechanism is related to the mRNA structure that can be different according to the mutation. The mRNA structure determines its stability, altering translation speed and protein folding.22
Haplotype analyses provide more complete information in association studies.16 In our work, different haplotype combinations were observed between males and females. In males, there was a statistically significant difference in the global distribution of haplotypes between the control group and the group of patients with both autoimmune and atopic disease. The haplotype H8 was only detected in patients (statistical power for this sample size 70%); whereas H1 was identified only in controls (statistical power 82%). These results again highlight that in males the group of patients that simultaneously developed both disorders seems to shape a well-defined group with a genetic background that appears to be unique to this clinical subgroup and is not seen in the groups of patients that have only one of the diseases.
The group of children that simultaneously present both immunological disorders is uncommon and there can be difficulties in finding sufficient subjects. For this reason we calculated the statistical power for our sample size and only associations with statistical significance higher than 70% are shown. Considering that 80% should be used, these results should be confirmed in larger populations although, as mentioned above, the group of children that simultaneously present both immunological disorders is uncommon. Another possible limitation of this study is the probability of reporting a false positive result due to multiple comparisons. Because of this, we calculated in all cases the probability of reporting a false positive result and only when this probability was less than 10% were the results considered. In addition, only associations with a significant p level less than 0.01 were considered instead of the 0.05.
It has been speculated that mutations in FOXP3 could be involved in each of the immunological pathologies included in this study (DM1, AT and CD). Several studies have examined the association of FOXP3 polymorphisms with these diseases and only one of our SNPs was reported. The 2680G>A polymorphism was identified in a study of rhinitis in an adult Chinese population but no association was observed.23 We did not find a relationship in our population either. In addition several studies analysed the FOXP3 gene in diabetic populations but only two found associations with some polymorphisms.24–29 No association has been found between FOXP3 polymorphisms and AT30,31 or CD.24,32
The FOXP3 polymorphisms have been analysed in patients with allergy in several studies and all of them found a statistical association.23,33–38 However, only five of them included children,24,28,29,33,34 with one of them including a Spanish population32 although none of them studied the entire coding region of FOXP3. In the present study we found an association between FOXP3 polymorphisms and the simultaneous development of allergic and autoimmune disorders.
To the best of our knowledge this is the first study to identify an association between FOXP3 polymorphisms in patients simultaneously suffering from Th1 and Th2 related disorders, suggesting that the genetic background of the FOXP3 gene may be involved in the development of several immunological diseases in the same patient, as occurs in the IPEX syndrome.2 Furthermore, these FOXP3 modifications could be involved in regulatory mechanisms implicated in these diseases. The critical role of FOXP3 in regulatory T cells by controlling the development of autoimmune and allergic diseases is clear,4,6 therefore FOXP3 mutations could be involved in the development of these diseases. The majority of SNPs detected in this study were located in regulatory regions. In fact, some authors have reported several mutations in the non-coding regions of FOXP3, such as UTR in patients with IPEX syndrome2,19 and others have confirmed epigenetic controls, such as histone methylation and acetylation, thus regulating transcription factor binding to the FOXP3 promoter region.4,5
To summarise, we have identified novel genetic variants of FOXP3 that are specifically present in children that simultaneously suffer from Th2 and Th1 disorders, atopic and autoimmune disease located mainly in regulatory regions. These variants could affect the expression levels of FOXP3, modifying its function. Further studies that analyse other regulatory sequences in this gene, as well as functional analysis, are required to better explain the role that FOXP3 polymorphisms play in these immunological disorders.
Conflict of interestThe authors have no conflict of interest to declare.
Ethical disclosuresPatients’ data protectionConfidentiality of data. The authors declare that no patient data appears in this article.
Right to privacy and informed consentThe authors declare that no patient data appears in this article.
Protection of human subjects and animals in researchThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
We thank the Allergy and Paediatric Departments of the University Hospital of Salamanca for helping in the participant recruitment. We are grateful to the colleagues of Allergy Laboratory for their help in lab procedures. We also express our gratitude to Prof. Innes Asher, Dr. Cameron Grant and Dr. Catherine Gilchrist for critical feedback on the manuscript.