To investigate whether genetic polymorphisms of C11orf30-LRRC32 region are associated with the development of childhood asthma in the Chinese population.
MethodsA total of 732 asthma children and 824 age-matched healthy controls were included in the study. Blood samples were collected from the subjects for total IgE analysis, DNA extraction and RNA extraction. Three previously reported asthma-related SNPs were genotyped, including rs7936070 (G/T), rs7927894 (A/G), and rs6592657 (A/G). Blood samples from 50 patients and 50 controls were randomly selected to detect the mRNA expression levels of C11orf30 and LRRC32 in serum.
ResultsThere were significantly different genotype frequencies between the two groups in terms of rs7936070 and rs7927894. Compared with controls, patients were found to have remarkably higher risk allele frequencies of rs7936070 and rs7927894. Genotype GG of rs7936070 was indicative of remarkably elevated total IgE level as compared with genotype TT and genotype GT. Similarly, genotype AA of rs7927894 was also associated with significantly elevated total IgE level. The serum expression of C11orf30 was significantly lower in the patients than in the controls. The C11orf30 expression was significantly correlated with the total IgE level (r = −0.463, p = 0.01).
ConclusionsVariants of C11orf30 were associated with the risk of childhood asthma in the Chinese population. Besides, abnormally decreased expression of C11orf30 was detected in the serum of patients, which was correlated with the total IgE level. The C11orf30 might play a role in asthma via biological pathways involving the regulation of total serum IgE level.
Asthma is the most frequent chronic respiratory disease that occurs during childhood.1 Characterized by airway epithelial damage and irreversible airway remodeling, asthma is associated with a high incidence of children morbidity and hospitalization.2,3 It is now accepted that asthma is a complex disease with heterogeneous phenotypes as well as variable degrees of atopic involvement.4 As shown by previous familial studies, genetic determinants may play a critical role in the development of asthma.5,6 It was estimated that genetic heritability may contribute to over 50 % of the total asthma risk.7–9 Herein, investigation of the genetic architecture of asthma can potentially improve the understanding of its etiology and thus contribute to the treatment of the disease.
In the last decade, genome-wide association studies (GWASs) have been used to uncover the susceptible genes of asthma.10–14 Moffatt et al.15 performed the first GWAS in 994 patients with childhood onset asthma and 1243 non-asthmatics. They found that SNPs in the ORMDL3-GSDMB region were significantly associated with the risk of childhood asthma in German and British populations.15 To date, over fifty susceptible loci of asthma have been uncovered through GWASs, such as PDE4D, RAD50-IL13, TLE4 and so on.16–18 To be noted, most GWASs have been performed in populations from European ancestry. Obviously, there was a lack of validation of these ethnic-specific loci in other populations.
In a recently published large GWAS, Zhu et al.19 analyzed data of 33,593 cases with asthma and allergic diseases from the UK Biobank. They found a strong genome-wide genetic correlation between asthma and variants located in the C11orf30-LRRC32 region.19 Interestingly, C11orf30 was previously reported as a genetic risk factor for food allergy by an independent GWAS performed in white population.20 In a subsequently replication study, C11orf30 was found to be strongly associated with total serum IgE levels.21 To the best of our knowledge, the association between asthma and the C11orf30-LRRC32 region remains uncertain in the Chinese population. In this study, we studied whether the genetic polymorphisms of the C11orf30-LRRC32 region are associated with the development of childhood asthma in the Chinese population.
MethodsSubjectsThe current case-control study was approved by the local institutional review board. A total of 1059 asthma children who received treatment from January 2009 to April 2018 in our clinic center were retrospectively reviewed. The diagnosis of asthma was confirmed according to the American Thoracic Society guidelines: 1. with a history of chest tightness and wheezing during the past year; 2. with a reversibility of forced expiratory volume in one second (FEV1) greater than 12 % spontaneously or after inhalation of b2-agonist. The inclusion criteria were as follows: 1. aged between 8 and 16 years; 2. with positive responses to a skin prick test (SPT; Allergopharma, Hamburg, Germany) using a panel of common allergens. Patients with other chronic inflammatory diseases or a history of treatment within three weeks before coming to our clinic were excluded from the study. Finally, 732 patients were included in the study. 824 age-matched healthy controls were recruited through routine physical examination. All the controls were excluded from having a history of respiratory disease or a familial history of asthma. The baseline characteristics of the subjects were recorded from the medical records, including age, gender, asthma severity, level of IgE and FEV1 %. Asthma severity was classified as mild (FEV1 %, >80 %), moderate (FEV1 %, 60–80 %), or severe (FEV1 % <60 %).
Collection of blood samplesAll the subjects gave their informed consent for the collection of the blood samples which were subsequently stored at −70 °C. The blood sample was used for total IgE analysis, DNA extraction and RNA extraction. Age-adjusted serum total IgE level was evaluated for each participant. Genomic DNA was extracted with a commercial kit (Qiagen K.K., Tokyo, Japan) following the standard protocol.
Genotyping of target variantsTaqMan Genotyping Assay was performed to detect the genotype of target variants. Three previously reported SNPs were genotyped, including rs7936070 (G/T), rs7927894 (A/G), and rs6592657 (A/G). ABI Prism Step-One-Plus Sequence Detection System (Applied Biosystems, Foster City, CA, USA) was used to analyze the outcome of the genotyping assay.
qPCRBlood samples from 50 patients and 50 controls were randomly selected to detect the mRNA expression levels of C11orf30 and LRRC32 in serum. Total RNA was extracted from the serum and then reversely transcribed in cDNA with a QuantiTect Reverse Transcription Kit (Qiagen, Tokyo, Japan). The used primers were as follows: forward 5′- GAAGTTCGGAGAGCAGTAAACG -3′, reverse 5′- ATCGACGACCTTCAATGGACC -3′ for C11orf30; forward 5′- AGACCCTTGATCTATCTGGGAAC -3′, reverse 5′- GAAGCTGATCTCATTGGTGCT -3′ for LRRC32; and forward 5′- AGACCCTTGATCTATCTGGGAAC -3′, reverse 5′- GAAGCTGATCTCATTGGTGCT -3′ for the endogenous control gene 18S. Cycle parameters of amplifying procedures were as follows, 94 °C for 10 min, followed by 36 cycles (94 °C for 15 s, 68 °C for 20 s, and 72 °C for 10 s) and finally 72 °C for 2 min. All samples were analyzed in triplicate.
Statistical analysisBaseline characteristics were compared between the patients and the controls by the Student-t test. The Hardy-Weinberg equilibrium (HWE) test was performed by the goodness-of-fit Chi-square test. The proportions of genotype and allele frequencies were compared between the two groups by the Chi-square test. Odds ratio (OR) was calculated for each SNP with the risk allele used as reference. One-way ANOVA test was used to analyze the relationship between genotypes and total IgE of the patients. Serum expression of C11orf30 and LRRC32 were compared between the cases and the controls using the Student-t test. The correlation between gene expression and clinical features was analyzed with the Pearson correlation analysis. All the data were analyzed with SPSS version 20.0 (SPSS Inc., Chicago, IL, USA). A P value of <0.05 was considered to be statistically significant.
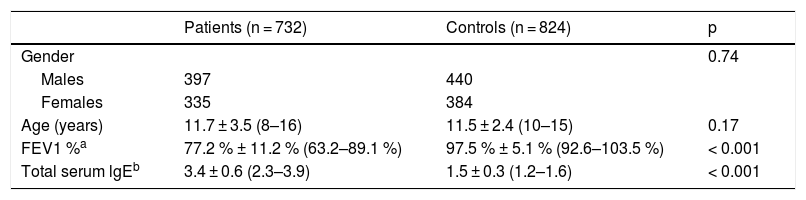
ResultsBaseline characteristicsThere were 397 male and 335 female patients with a mean age of 11.7 ± 3.5 years. As summarized in Table 1, there was no significant difference between the cases and the controls regarding age and the ratio of males to females. Patients were found to have remarkably higher level of total serum lgE than the controls (3.4 ± 0.6 vs. 1.5 ± 0.3, p < 0.001). Decreased pulmonary function was observed in the patients, with a mean FEV1 of 77.2 % ± 11.2 %. According to FEV1 %, 395 (54.0 %) patients were classified as mild asthma, 180 (24.6 %) as moderate and 157 (21.4 %) as severe asthma.
Demographic data of the participants.
| Patients (n = 732) | Controls (n = 824) | p | |
|---|---|---|---|
| Gender | 0.74 | ||
| Males | 397 | 440 | |
| Females | 335 | 384 | |
| Age (years) | 11.7 ± 3.5 (8–16) | 11.5 ± 2.4 (10–15) | 0.17 |
| FEV1 %a | 77.2 % ± 11.2 % (63.2–89.1 %) | 97.5 % ± 5.1 % (92.6–103.5 %) | < 0.001 |
| Total serum lgEb | 3.4 ± 0.6 (2.3–3.9) | 1.5 ± 0.3 (1.2–1.6) | < 0.001 |
NS, not significant; FEV1, forced expiratory volume in the first second of expiration.
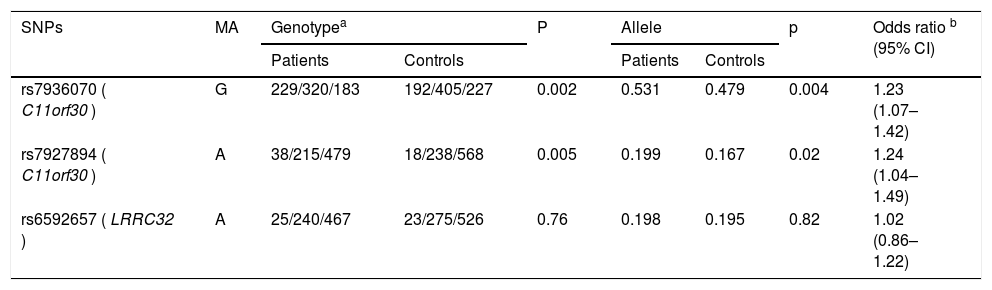
Distributions of genotype and allele frequency of the three genotyped variants are summarized in Table 2. As shown by the HWE test, there was no remarkable deviation regarding the genotype frequency in the controls. There were significantly different genotype frequencies between the two groups in terms of rs7936070 and rs7927894 (31.3 % vs. 23.3 %, p = 0.002 for genotype GG of rs7936070; 5.2 % vs. 2.2 %, p = 0.005 for genotype AA of rs7927894). Compared with the controls, patients were found to have remarkably higher risk allele frequencies of rs7936070 and rs7927894 (53.1 % vs. 47.9 %, p = 0.004 for rs7936070; 19.9 % vs. 16.7 %, p = 0.02 for rs7927894), with an OR of 1.23 and 1.24, respectively. As for rs6592657, there was no significant difference between the patients and the controls regarding the frequency of allele and genotype.
Association of the three loci with development of asthma.
| SNPs | MA | Genotypea | P | Allele | p | Odds ratio b (95% CI) | ||
|---|---|---|---|---|---|---|---|---|
| Patients | Controls | Patients | Controls | |||||
| rs7936070 ( C11orf30 ) | G | 229/320/183 | 192/405/227 | 0.002 | 0.531 | 0.479 | 0.004 | 1.23 (1.07–1.42) |
| rs7927894 ( C11orf30 ) | A | 38/215/479 | 18/238/568 | 0.005 | 0.199 | 0.167 | 0.02 | 1.24 (1.04–1.49) |
| rs6592657 ( LRRC32 ) | A | 25/240/467 | 23/275/526 | 0.76 | 0.198 | 0.195 | 0.82 | 1.02 (0.86–1.22) |
NS, not significant; CI, confidential interval; MA, minor allele.
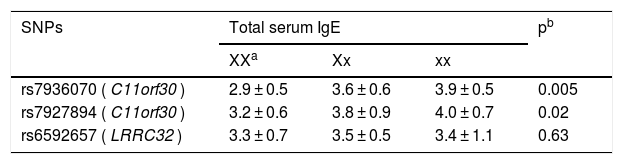
As shown in Table 3, genotype GG of rs7936070 was indicative of remarkably elevated total IgE level (3.9 ± 0.5) as compared with genotype TT (2.9 ± 0.5) and genotype GT (3.6 ± 0.6). Similarly, genotype AA of rs7927894 was also associated with significantly elevated total IgE level (4.0 ± 0.7). As for rs6592657, no significant association with the serum IgE level was found.
Serum expression of C11orf30 and LRRC32The serum expression of C11orf30 was significantly lower in the patients than in the controls (0.000022 ± 0.000013 vs. 0.000037 ± 0.000022, p < 0.001). The C11orf30 expression was significantly correlated with the total IgE level (r = −0.463, p = 0.01). As for LRRC32, no significant difference of the mean expression was found between the two groups (0.0000029 ± 0.0000034 vs. 0.0000031 ± 0.0000024, p = 0.73). There was no obvious correlation between the LRRC32 expression and the total IgE level (r = −0.155, p = 0.62).
DiscussionEarlier literature has implied the potential role of the C11orf30-LRRC32 region in the development of inflammatory disease.19–22 In a published GWAS, rs7927894 located between the C11orf30 and LRRC32 genes was firstly identified as a susceptible locus of atopic dermatitis.23 O’Regan et al.24 further confirmed that rs7927894 was associated with childhood eczema. In another replication study, Li et al.21 studied the association of the C11orf30-LRRC32 region with total serum IgE levels of asthma patients in three independently ascertained white populations. They found four SNPs in this region significantly associated with total serum IgE levels, including rs7130588, rs2508746, rs10899234, and rs6592657.21 Of note, rs7130588, rs2508746 and rs10899234 were all in linkage disequilibrium (LD) with rs7927894.21 As shown by the GWAS performed by Zhu et al.,19 variant rs7936070, located in the C11orf30-LRRC32 region, was remarkably associated with asthma. In this study, we validated that two SNPs of the C11orf30-LRRC32 region, rs7936070 and rs7927894, are associated with the risk of childhood asthma in the Chinese population. Allele G of rs7936070 and allele A of rs7927894 could add to the risk of childhood asthma 1.23 fold and 1.24 fold, respectively. Comparably, Zhu et al.19 reported an OR of 1.13 for allele G of rs7936070 in patients with asthma.
To further characterize the role of rs7936070 and rs7927894 in childhood asthma, we investigated the relationship between risk alleles and total serum IgE levels of the patients. In line with the finding of Li et al.,21 genotype AA of rs7927894 was remarkably associated elevated IgE level in our study. In addition, genotype GG of rs7936070 was also found indicative of significantly higher IgE level. Obviously, the association between the C11orf30-LRRC32 region and total serum IgE levels existed in populations other than non-Hispanic whites.
To determine the causative gene of asthma in the C11orf30-LRRC32 region, serum expression of these two genes was detected in the current study. Significantly decreased expression of C11orf30 was found in the patients as compared with the controls. Interestingly, the C11orf30 expression was significantly correlated with the total IgE level. By contrast, there was no significant difference between the two groups regarding the expression of LRRC32, the expression level of which was not correlated with the total IgE level either. Based on these findings, we speculated that C11orf30 could be the real causative gene of asthma.
The biological function of C11orf30 remains obscure. It encodes the EMSY protein which may play a role in the function of epithelial barrier.23C11orf30 was also reported as a risk factor of polysensitization to allergens.22 It was hypothesized that a dysfunctional epithelial barrier could be associated with high sensitization implicated in atopic asthma.22 As shown by the ENCODE database, rs7936070 is located in possible enhancer sites and thus can potentially influence the expression level of C11orf30.25 Further in-vivo cellular experiment investigating the relationship between rs7936070 and C11orf30 expression is warranted.
Although rs6592657 was reported to be associated with total serum IgE level of asthma patients, we failed to validate the association of this variant with asthma. Located in the intron of LRRC32, rs6592657 was not in LD with rs7936070 or rs7927894. It partially indicated that LRRC32 might not be involved in the development of asthma in the Chinese population. It was previously reported that genetic associations for asthma varied among different ethnic groups. Therefore, the inconsistence between our findings and previously reported results might be attributed to the ethnic heterogeneity. In future studies, variants of LRRC32 should be validated in other populations to determine its role in asthma.
As an inherent limitation of candidate gene association study, the statistical power to detect significant association is scarce. In the current study, evidence of replication in Asians was found for the locus previously identified in Europeans, suggesting its role in asthma of different populations. In a future study, a genome-wide approach in the Asian population can provide new insights into the genetic architecture of asthma.
ConclusionsThe relationship between the C11orf30-LRRC32 region and asthma was refined in the current study. Variants of C11orf30 were found associated with the risk of childhood asthma in the Chinese population. Besides, abnormally decreased expression of C11orf30 was detected in the serum of patients, which was correlated with the total IgE level. The C11orf30 might play a role in asthma via biological pathways involving the regulation of total serum IgE level. The functional role of currently validated susceptible variants needs to be further explored.
Conflict of interestThe authors involved in this study do not have anything to disclose regarding funding or any conflicts of interest with respect to this manuscript.
Ethical disclosuresThe authors declare that all the patients included in the study have received sufficient information and have given their informed consent in writing to participate in this study.