INTRODUCTION
Coeliac disease (CD) shows a complex pattern of heredity. Predisposition to the disease might be partially due to an individual pattern of immune response with a trend to hyper-inflammatory bias and an excessed production of proinflammatory factors. This phenotype may represent an evolutionary advantage in an environment dominated by infectious agents. One of these patterns of intense response is codified within the Ancestral Haplotype (AH) 8.1 of HLA, shared by a great number of CD patients and patients with other autoimmune diseases. This AH carries the main risk alleles known for CD: those that codify for the HLA-DQ2 heterodimer (DQB1*02 and DQA1*0501), and allele A (or 2) of the biallelic polymorphism in position 308 of the TNF gene.
The DQ2 heterodimer plays a defined role in the pathogenesis of CD, acting as a restriction factor for the presentation of gluten peptides to T lymphocytes after processing by antigen presenting cells (APCs). It has been previously discussed the role of the TNF gene as an independent risk factor for CD. However, 10 % of CD patients do not expressed the DQ2 heterodimer: most of them express the DQ8 heterodimer, with a similar ability to link gluten peptides than DQ2, or other heterodimers which share one of the chains with DQ2, but they usually do not carry the TNF 308A allele. Furthermore, many of the CD patients in the south of Europe codify the α/β chains of the DQ2 heterodimer in trans, within a non-AH 8.1 haplotype, and they do not carry TNF 308A. Carriers of this allele associate to an increased production in vitro of TNFα by mononuclear cells. A similar response might be achieved by the intervention of other genes controlling the expression of other proinflammatory or antiinflammatory cytokines as IL6, IL10 and TGFβ1.
The aims of this work are to study the frequency of polymorphisms of several genes with a relevant function on the production of TNFα, IL10, IL6 and TGFβ1 in DQ2 negative CD patients, and to compare the results with DQ2 positive patients and healthy controls, in order to establish whether any of these polymorphisms has a role in CD susceptibility.
PATIENTS AND METHODS
We tested genomic DNA samples of 51 CD patients (27 DQ2 + ve and 24 DQ2-ve) and 99 healthy controls. Samples from CD patients were collected from the Paediatric Gastroenterology Clinics at the Hospital Clínico Universitario of Valladolid and Hospital Sant Joan de Déu of Barcelona. Control samples were collected from blood donors attending to the Blood Bank of the Valladolid district. Barcelona city has a mixed population coming from other regions of Spain, so it is not expected a great genetic bias from population of the center of the country.
All CD patients had compatible symptoms of the disease, positive serology (IgA antiendomysial or antitransglutaminase antibodies), and mucosal changes in the small intestinal biopsy at the time of diagnosis, as well as signs of clinical, serologic and pathological recovery after gluten free diet. They were previously typed for HLA-DQ2 and grouped into DQ2 positive and DQ2 negative CD patients. Healthy controls were also HLA-DQ2 typed.
DQ2 positive samples were defined as having both risk alleles of CD: HLA-DQB1*02 and HLA-DQA1*0501. Specific DQ2 alleles were determined as previously described.
TNF 308 (G > A), IL-6 174 (G > C) and TGFB codon 10 (+ 869, T > C) and codon 25 (+ 915, G > C) polymorphisms and IL-10 haplotype of polymorphisms in positions 1082 (G > A), 819 (C > T) and 592 (C > A) were typed by a SSP-PCR technique (Cytokine Genotyping Tray, One Lambda Inc. CA, USA). We compared TGFB1, IL-6, allele, allele carrier and genotype frequencies and IL-10 haplotype, genotype, allele and allele carrier frequencies amongst DQ2 positive and negative CD patients, and healthy controls, and also with the presence of allele A (2) of TNF 308 (ie. high producers of TNFα).
Statistical analysis was performed by using the SPSS v11.0 software. Contingency tables and the Chi square of Pearson, with asintotic significance (p) and Fisher's exact correction in 2 x 2 tables were calculated. Risk was expressed as Odd Ratio (OR) and 95 % Confidence Interval [CI].
RESULTS
Cytokine gene polymorphisms in DQ2 positive CD patients
The distribution of allele frequencies for TNF 308 (table I), is different between DQ2 positive CD patients and controls and the same occurs for the haplotype frequencies of the IL10 promoter (1082, 819, 592) (p = 0.027; table II). The frequencies of the TNF 308A allele (p = 0.027), TNF 308A carriers (p = 0.031) and of the IL10GCC haplotype, are increased compared to the remainder haplotypes (48.0 % vs. 29.9 %, p = 0.013, OR = 2.15 [1.15-4.08]). In contrast, the frequency of IL10ATA haplotype vs. the remainder haplopytes is decreased in DQ2 positive patients as compared to healthy controls (23.1 % vs. 38.7 %, p = 0.025, OR = 0.47 [0.23-0.96]), or DQ2 positive controls (23.1 % vs. 42.2, p = 0.036, OR = 0.41 [0.17-0.98]).
When DQ2 positive CD patients and healthy controls were stratified in TNF 308A carriers and non carriers, we observed an increased frequency of the allele C of codon 25 of the TGFB1 gene (41.6 % vs. 15.5 %, p = 0.014) in CD patients non carriers of TNF*A, and an increase of homozygous CC genotype of IL6 174 in patients carriers of TNF 308A (50 % vs. 11 %, p = 0.042).
Cytokine gene polymorphisms in DQ2 negative CD patients
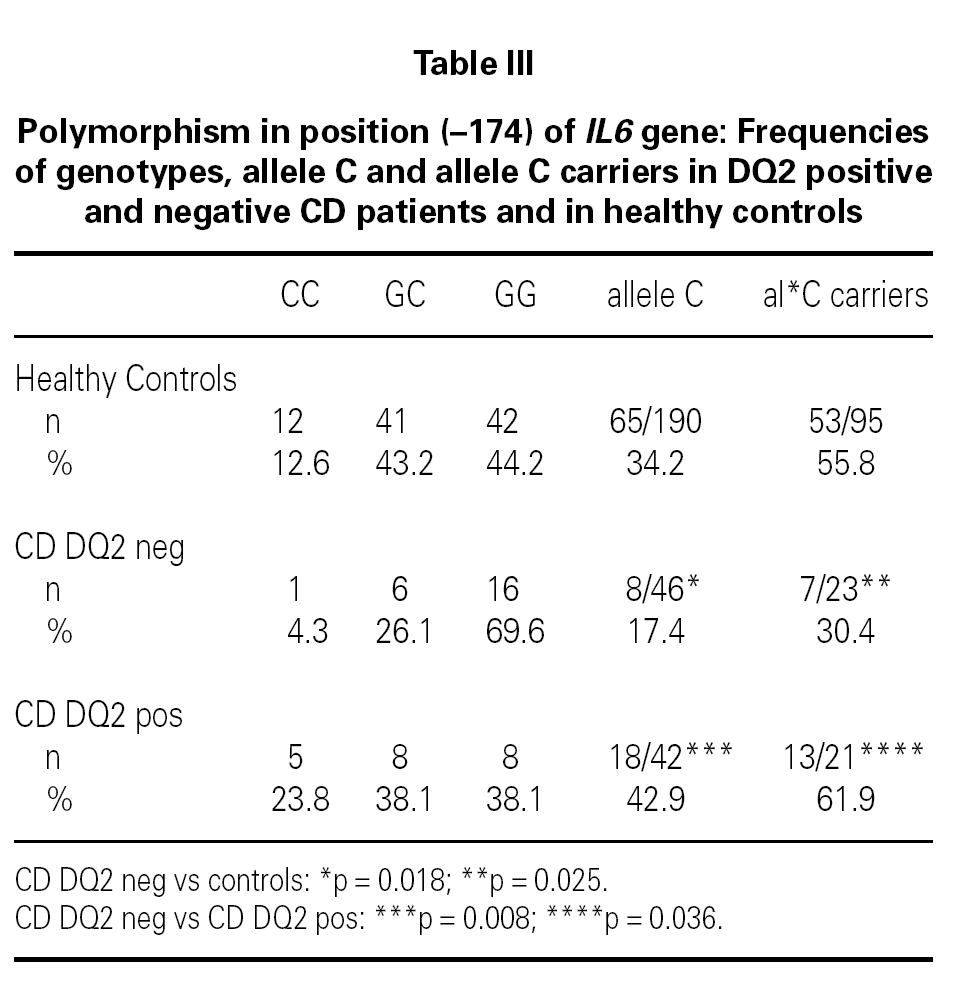
The IL6 174 allele G is more frequent in DQ2 negative patients than in controls (p = 0.018, OR = 2.47 [1.09-5.60]; table III) or DQ2 negative controls (p = 0,018, OR = 2.60 [1.11-6.09]; table III). There is also an increased frequency of non carriers of the allele C of IL6 174 in DQ2 negative CD patients when compared with controls (p = 0.025, OR = 2.88 [1.09-7.66]; table III), with DQ2 negative controls (69.6 % vs. 44.8 %, p = 0.038, OR = 2.81 [1.01-7.86]) and with DQ2 positive CD patients (p = 0.036; table III).
No differences were found between DQ2 negative CD patients and healthy controls in the frequency distribution of TNF 308A carriers. Stratifying these groups as TNF 308A carriers and non carriers, we found that 76.5 % of TNF 308A non carriers-DQ2 negative CD patients are IL6 174C non carriers (IL6 174GG homozygous), as compared to 43.5 % of TNF 308A non carriers-healthy controls (p = 0.014, OR = 4.22[1.15-14.28]).
Differences between DQ2 positive and negative CD patients
There is an increased frequency of the TNF 308A allele within the group of DQ2 positive CD patients compared to DQ2 negative cases (p = 0.031; table I), while DQ2 negative patients show a higher frequency of the IL6 174G allele (table III; p = 0.008) and of IL10 1082A, although without reaching statistical significance (66.7 % vs. 47.8 %, p = 0.065). Non carriers of the allele C of IL6 174 (IL6 174GG homozygous) are more frequent amongst DQ2 negative patients (p = 0.036; table III).
No differences have been found in the frequency distribution of frequencies of alleles, genotypes or haplotypes of TGFB1 (codons 10 and 25) (table IV). However, differences are observed in haplotypes frequencies of the IL10 gene promoter although they do not reach statistical significance (table II).
DISCUSSION
We have studied cytokine gene biallelic polymorphisms in 2 different groups of CD patients and in a group of ethnically matched healthy controls. We have typed a group of DQ2 negative CD patients and a similar sample size of DQ2 positive patients. The polymorphisms studied had been described as having a functional correlate in an in vitro production of the corresponding cytokine. We had previously studied the value of allele 2 (A) in position 308 of the promoter of TNF and allele 1 (G) of NcoI RFLP in 1st intron of LTA gene as a putative additional risk markers for CD within DQ2 positive patients. Here, we have assessed the role of IL6 174, IL10 (1082, 819 and 592) promoter gene polymorphisms and TGFB1 codon 10 and codon 25 polymorphisms in CD susceptibility. HLA-DQ, IL6, IL10 and TGFB1 loci are in different chromosomes, and they segregate independently.
It is known that diseases with a polygenic pattern of heredity, as CD, are manifested when a threshold of susceptibility burden is overcrossed, by acummulation of risk factors. HLA-DQ contributes to up to 40 % of the genetic component of this burden and it is the best risk factor known for CD disease. However, other factors should be involved, and perhaps they are different amongst several groups of patients and/or populations. Genes codifying for several molecules related to the immune system response have been implicated in CD susceptibility: CTLA4, MICA and MICB or HSP70. Evidence of IFNγ gene involvement in CD susceptibility have been reported within a Spanish population by means of family transmission studies. The patterns of IL10-1082 allele A associated TNF 308 allele A was described in a group of Italian CD patients with IgA deficiency.
We have found differences in the proinflammatory factor genes between CD patient groups: TNF gene involvement is more frequent in DQ2 positve CD patients, with high frequencies of genotypes linked to TNFα high producer phenotype, in agreement to previous reports. However, the IL6 high producer phenotype (IL6 174GG homozygous genotype) is more frequent amongst DQ2 negative patients, as compared to healthy controls or DQ2 positive patients. Furthermore, the IL6 174GG homozygous genotype is increased amongst TNF 308A non carrier CD patients as compared to heathy controls (p = 0.023, OR = 2.66 [1.09-6.49]) and TNF 308A non carrier controls (p = 0.025, OR = 2.74 [1.09-6.92]) data not shown. Recently Woolley reported no evidence of association between TGF-beta1, IL-10, IL-6 and IFN-gamma polymorphisms and CD susceptibility in a Finnish population. The genetic heterogenicity of Spanish CD patients may explain the differences of the results with Finnish CD patients, where most of the DQ2 positive CD patients present AH 8.1, versus more DQ2 in trans, non carriers of TNF 308A, in Spanish ones and also there are genetic differences in the DQ2 negative patients.
The involvement of the IL10 gene is less clear. No differences were found in genotype and allele frequencies between the two groups of CD patients or when CD groups were compared to controls. However, an increased frequency of the GCC haplotype (associated to high production of IL10) is observed in DQ2 positive patients compared to healthy controls, whereas the ATA haplotype may be a protective marker for these group of patients. In contrast, a subgroup of DQ2 negative patients, TNF 308A non-carriers (TNFα low producer phenotype), show an increased frequency of IL10 1082A carriers (low IL10 producers). These findings are different to other reports in CD patients with IgA deficiency, although none of our patients had IgA deficiency.
TGFβ is an important regulatory molecule, but the TGFB1 gene does not seem to have any relation with CD suceptibility in our population. We have found a slightly increased allele C carriers of the polymorphism in codon 25 in a small subgroup of DQ2 positive patients (TNF 308A non carriers). The allele C in codon 25 polymorphism is associated with intermediate-low production of TGFβ1. However, this allele is the most frequent within the healthy population, and our results, although statistically significant, might be an artefact due to the low number of cases.
In conclusion, DQ2 negative CD patients show an increased frequency of genotypes of IL6 high producers. These were mainly allele G homozygous for the IL6 gene (174) polymorphism. The IL6 174GG genotype (homozygous) may be an additional risk marker for CD in DQ2 negative patients, representing an alternative susceptibility factor for CD when TNF 308A is negative, although a wider population study is needed to validate these findings.
ACKNOWLEDGMENTS
This work was supported by grants from the Instituto de Salud Carlos III, of the Spanish Ministry of Health (PI020895, 02/3068), Junta de Castilla y León (VA057/04), and Sweden Diagnostics affiliated to Pharmacia Diagnostics.