Considering the possible roles of interleukin-23 receptor (IL-23R) gene in the pathogenesis of juvenile systemic lupus erythematosus (JSLE), the objective of this study was to elucidate whether polymorphisms of the IL23R are associated with susceptibility to JSLE in an Iranian population.
Materials and methodsA case-control study on 62 patients with JSLE and 78 healthy controls was performed to investigate the associations of four single nucleotide polymorphisms (SNPs) in IL-23R gene, namely, rs7517847, rs10489629, rs11209026, and rs1343151, with susceptibility to JSLE, using real-time polymerase chain reaction Taqman genotyping technique.
ResultsAnalysis of allele and genotype frequency of four selected SNPs revealed statistically significant positive association between homozygous variant of rs7517847 (TT) (P, 0.02) and T allele at the same position (P, 0.01) with JSLE vulnerability. There was no significant association between other evaluated SNPs and JSLE susceptibility.
ConclusionThese findings suggest that particular IL-23R gene variants could affect individual susceptibility to JSLE.
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disorder with diverse clinical manifestations and unclear pathogenesis. The disease is associated with the production of autoantibodies against nuclear and cytoplasmic components, abnormalities of immune-inflammatory system functions and inflammatory manifestations in several organs.1 Juvenile systemic lupus erythematosus (JSLE), constitutes 10–20% of all SLE cases and predominantly appears before the age of 16 years.2,3 Although JSLE and adult-onset SLE share several features, JSLE is found to be more severe than the adult-onset SLE, leading to higher incidence of organ involvement and more rapid clinical progression.4 A complex interplay between genetic risk factors and environmental events is hypothesized to contribute towards disease initiation and progression.3,5 Genome-wide association studies have revealed familial tendency for disease expression, such as the evident higher concordance of SLE in monozygotic twins in comparison to healthy subjects.1,6 The results of such studies together with the data provided by our previous studies have supported the notion that genetic factors such as the genes coding for cytokines and their receptors could play crucial roles in the pathogenesis of inflammatory disorders such as JSLE.7–18
Interleukin-23 (IL-23) is a heterodimeric cytokine composed of a unique p19 subunit and a common p40 subunit shared with IL-12.19 IL-23 is mainly expressed through activation of phagocytes and dendritic cells20,21 and has been shown to play an essential role in the activation of a subset of CD4+T cells characterized by the production of the interleukin-17 (IL-17), namely, Th17 cells.22,23 IL-17 induces the synthesis of proinflammatory cytokines which ultimately leads to chronic inflammation and the destruction of joints.21,24 All these findings suggest IL-23 pathway as an important actor in the pathogenesis of chronic inflammatory diseases such as SLE.22 IL-23 exerts its effects through binding to the IL-23 receptor (IL-23R) complex with high affinity.20 IL-23R constitutes a common IL-23 receptor and an IL-12 receptor β1 subunit, which are mostly expressed on activated and memory T cells.22,25 The IL-23R gene is positioned on chromosome 1p31, spanning 2.8kb and comprising 11 exons and 10 introns. The promoter region of the human IL-23R gene is distributed with many single nucleotide polymorphisms.26 A multitude of studies have depicted a strong association between IL-23R polymorphisms and the progression and outcome of several autoimmune disorders, such as ankylosing spondylitis, Crohn’s disease, and rheumatoid arthritis.21 The aim of this study was to investigate the role of IL23R variants in JSLE susceptibility. Herein, we have examined the association between four selected IL-23R polymorphisms and JSLE proneness in Iranian population.
Materials and methodsThe study population included 62 consecutive Iranian patients with JSLE (with a mean age of 11 years and range of 4–14 years), who were referred to the Rheumatology Clinic of the Children’s Medical Center Hospital, the pediatrics center of excellence in Iran, between March 2015 and April 2018, and fulfilled the American College of Rheumatology (ACR) classification criteria for SLE.4 Thorough history taking, comprehensive examination and relevant laboratory and radiological studies were carried out for all patients. Patients diagnosed with any other concomitant disorder were excluded from the study. Seventy-eight ethnically, age and sex matched healthy controls with no family history or clinical manifestation of any rheumatologic and autoimmune disorders were also randomly recruited from those who visited for routine check-ups at the same center. All the patients and controls were unrelated individuals. Written informed consent was obtained from all the subjects’ parents or guardians according to the guidelines of the Ethics Committee of Tehran University of Medical Sciences before blood sampling.
Genomic DNA samples were obtained from all the participants and were stored at −20°C until administration. DNA samples of patients and healthy controls were extracted from peripheral blood leukocytes using the phenol-chloroform method. Genotyping of the samples was conducted by real-time polymerase chain reaction (RT-PCR), using TaqMan probe (ABI, USA) with an ABI 7300 RT-PCR instrument (Applied Biosystems). Optical 96-well reaction plate 0.2μl (ABI, USA) was administered for the test. The total volume of 20μl in each microwell, consisted of 10μl Master Mix (ABI, USA), 0.5μl Assay Mix (ABI, USA), 4.5μl deionized water, and 5μl DNA samples with a concentration of 20ng/ml. PCR conditions were as follows: 95°C for 10min, followed by 40 cycles of 95°C for 15s and 60°C for 1min. The results were determined using Allelic Discrimination Program (Applied Biosystems).
Genotype and allelic frequencies of certain IL-23R SNPs, including rs10489629, rs11209026, rs1343151, and rs7517847 (Table 1) were compared between JSLE cases and controls using the chi-square test and Fisher’s exact test when appropriate, and odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the relative risk conferred by a particular allele and genotype. Hardy-Weinberg equilibrium evaluation was performed for each polymorphism included in this research. All SNPs were found to be in Hardy–Weinberg equilibrium (HWE) (P>0.05) in the control group. Statistical significance was assumed at the p<0.05 level. The statistical program Epi Info 7 was used for all statistical analyses.
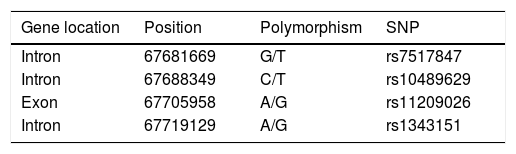
Details of IL-23R gene single nucleotide polymorphisms (SNPs), gene polymorphisms, positions and locations, assessed in the present investigation.
| Gene location | Position | Polymorphism | SNP |
|---|---|---|---|
| Intron | 67681669 | G/T | rs7517847 |
| Intron | 67688349 | C/T | rs10489629 |
| Exon | 67705958 | A/G | rs11209026 |
| Intron | 67719129 | A/G | rs1343151 |
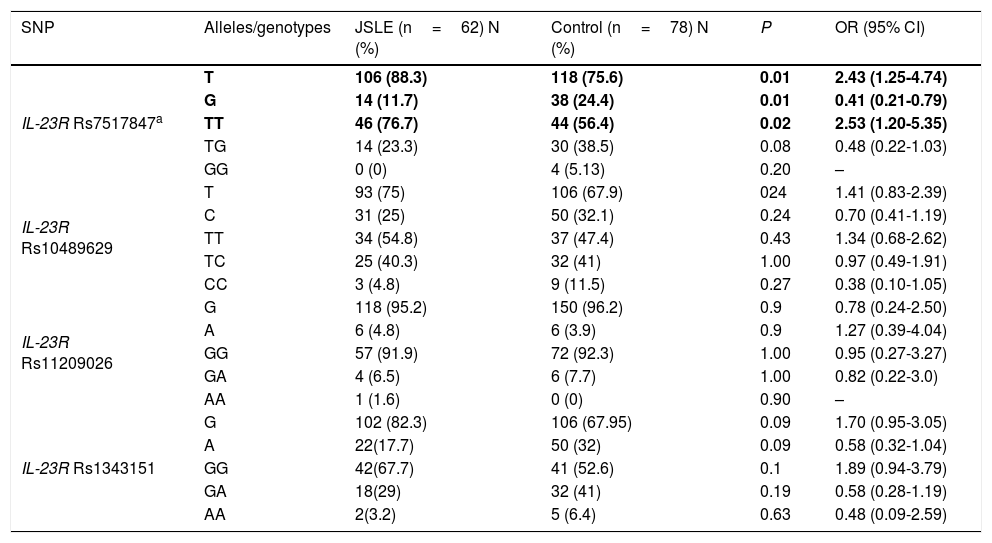
Allelic and genotype frequencies of rs7517847, rs10489629, rs11209026, and rs1343151 polymorphisms in patients with juvenile systemic lupus erythematosus and healthy control subjects are depicted in Table 2.
Allele and genotype distribution of IL-23R gene in JSLE patients and healthy controls.
| SNP | Alleles/genotypes | JSLE (n=62) N (%) | Control (n=78) N (%) | P | OR (95% CI) |
|---|---|---|---|---|---|
| IL-23R Rs7517847a | T | 106 (88.3) | 118 (75.6) | 0.01 | 2.43 (1.25-4.74) |
| G | 14 (11.7) | 38 (24.4) | 0.01 | 0.41 (0.21-0.79) | |
| TT | 46 (76.7) | 44 (56.4) | 0.02 | 2.53 (1.20-5.35) | |
| TG | 14 (23.3) | 30 (38.5) | 0.08 | 0.48 (0.22-1.03) | |
| GG | 0 (0) | 4 (5.13) | 0.20 | – | |
| IL-23R Rs10489629 | T | 93 (75) | 106 (67.9) | 024 | 1.41 (0.83-2.39) |
| C | 31 (25) | 50 (32.1) | 0.24 | 0.70 (0.41-1.19) | |
| TT | 34 (54.8) | 37 (47.4) | 0.43 | 1.34 (0.68-2.62) | |
| TC | 25 (40.3) | 32 (41) | 1.00 | 0.97 (0.49-1.91) | |
| CC | 3 (4.8) | 9 (11.5) | 0.27 | 0.38 (0.10-1.05) | |
| IL-23R Rs11209026 | G | 118 (95.2) | 150 (96.2) | 0.9 | 0.78 (0.24-2.50) |
| A | 6 (4.8) | 6 (3.9) | 0.9 | 1.27 (0.39-4.04) | |
| GG | 57 (91.9) | 72 (92.3) | 1.00 | 0.95 (0.27-3.27) | |
| GA | 4 (6.5) | 6 (7.7) | 1.00 | 0.82 (0.22-3.0) | |
| AA | 1 (1.6) | 0 (0) | 0.90 | – | |
| IL-23R Rs1343151 | G | 102 (82.3) | 106 (67.95) | 0.09 | 1.70 (0.95-3.05) |
| A | 22(17.7) | 50 (32) | 0.09 | 0.58 (0.32-1.04) | |
| GG | 42(67.7) | 41 (52.6) | 0.1 | 1.89 (0.94-3.79) | |
| GA | 18(29) | 32 (41) | 0.19 | 0.58 (0.28-1.19) | |
| AA | 2(3.2) | 5 (6.4) | 0.63 | 0.48 (0.09-2.59) |
JSLE: Juvenile systemic lupus erythematosus; OR: odds ratio; CI: confidence interval.
Analysis of allele and genotype frequency of four selected SNPs revealed a statistically significant positive association between the homozygous variant of rs7517847 (TT) (76.7% vs. 56.4%; P, 0.02; OR, 2.53; 95%CI, 1.20–5.35) and T allele at the same position (88.3% vs. 75.6%; P, 0.01; OR, 2.43; 95%CI, 1.25–4.74) with JSLE vulnerability in our study. There was no significant association between other evaluated SNPs and JSLE susceptibility.
DiscussionTo the best of our knowledge, the present investigation is the first to assess the correlation between IL-23R polymorphisms, which have been previously postulated as a possible genetic marker for autoimmunity,22 and JSLE in an Iranian population. Our results showed statistically significant correlation between both the IL-23R rs7517847T allele and TT genotype and individuals’ susceptibility to JSLE. On the other hand, we could not detect any significant difference between the case and control groups in the frequency of IL-23R rs11209026, rs1343151, and rs10489629 variants.
The results of the current investigation are not in line with the findings of the study conducted by Yi Li et al. demonstrating no significant differences in the genotype and allele frequencies of rs10889677, rs1884444, and rs7517847 polymorphisms between the patients with SLE and the control group in a Chinese population.27 In another study, Sanchez et al. evaluated eight IL-23R SNPs (rs1004819, rs7517847, rs10489629, rs11209026, rs1343151, rs10889677, rs11209032, and rs1495965) and revealed no statistically significant differences between SLE patients and healthy controls of Spanish origin.22 The results of these above-mentioned investigations are consistent with the findings of a study conducted by Kim et al. showing lack of association between seven IL-23R SNPs (rs1004819, rs7517847, rs10489629, rs2201841, rs1343151, rs11209032, and rs1495965) and SLE susceptibility in a sample of over 600 Korean SLE patients and almost 1000 healthy controls.28 Consistently, Safrany et al. observed no significant difference in the IL-23R rs11805303, rs10889677, rs1004819, rs2201841, rs11209032, 11209026, rs10489629, rs7517847, and rs7530511 variants between the SLE patient and control groups in the Hungarian population.29 In 2018, Imani et al.21 performed a systematic review and meta-analysis, pooling the results of all the four above-said studies, and depicted a significant association between the IL-23R gene rs7517847 T>G SNP and SLE risk, in line with our results. Our literature search also revealed another study,30 assessing IL-23R rs10889677, and rs1884444 SNPs, indicating lack of association of these polymorphisms with SLE susceptibility or severity. Notwithstanding the fact that our investigation implied rs7517847 variant of IL23R as a novel susceptibility gene in JSLE, the lack of association between certain IL-23R gene variants and SLE vulnerability found in a number of previous studies could possibly indicate that IL-23R may play an important role in regulating local inflammation rather than systemic inflammatory processes involved in systemic autoimmune disease such as systemic sclerosis.31,32 However, IL-23R gene was found to be associated with organ-characteristic autoimmune diseases such as inflammatory bowel disease,33 experimental autoimmune encephalomyelitis,34 rheumatoid arthritis,35 ankylosing spondylitis36 and psoriasis.37 This lack of association could also be partly attributed to the fact that IL-23 preferentially stimulates T cells to produce cytokines, including tumor necrosis factor-a, IL-6, and IL-17, but does not provoke the production of type I interferons, which are assumed to play a relevant role in the development and maintenance of the disease process in SLE.38,39
It should be noted that the current study has some certain constraints, including the relatively small number of participants in the patients’ category, resulting in diminished statistical power of the analysis; together with our limitation to measure serum levels of IL-23 and IL-17, which altogether limits the ability of our investigation to reach a consolidated answer to the questions considering the precise role of the above-mentioned gene variants in both the cytokines’ production and JSLE pathogenesis.
To conclude, the results of the current investigation suggested rs7517847 variant of IL23R as a novel susceptibility gene in JSLE. However, as this gene is not yet widely evaluated in JSLE patients, we are unable to incisively accept or reject the definite role of this gene in the pathogenesis of JSLE. Further multi-center studies enrolling larger sample sizes of different ethnicities are required to confirm this finding.
FundingThis study was supported by a grant from Tehran University of Medical Sciences (95-03-154-32285).
Conflict of interestIt should be noted that there is no ethical problem (approved by the research ethics committee of Tehran University of Medical Sciences) or conflict of interest in our research. There was no honorarium, grant, or other form of payment to authors to produce the manuscript.