Background. The first line anti-tubercular (anti-TB) treatment normally involves isoniazid, rifampicin, pyrazinamide, and ethambutol. Clearance of these drugs depends on the activity of several enzymes such as N-acetyl transferase 2, cytochrome P450 oxidase and glutathione S-transferase (GST). Some of these enzymes are highly polymorphic leading to significant inter-individual variation in their activity thereby increasing the risk of drug induced hepatotoxicity (DIH).
Aim. To investigate the possible association of anti-TB DIH with genetic polymorphism of GST genes in Western Indian population.
Material and methods. A prospective case-control study was undertaken on patients who received anti-TB treatment. Cases (n = 50) were distinguished from controls (n = 246) based on occurrence of DIH during anti-tubercular treatment. A multiplex polymerase chain reaction was employed to identify homozygous null mutation at GSTM1 and GSTT1 loci.
Results. Homozygous null mutation in GSTM1 gene alone or in both GSTM1 and T1 genes was found to be significantly associated with anti-TB DIH at p < 0.02 and p < 0.007, respectively, in our study population.
Conclusions. This is the first study to report GSTM1 null and combined GSTM1 and T1 null genotypes to be risk factors of anti-TB DIH in Western Indian population. Screening of patients for these genotypes prior to anti-TB regimen would provide better control of hepatotoxicity.
Tuberculosis (TB) is endemic in developing countries. According to a recent WHO report, India with 1.98 million cases, accounts for approximately 26% of TB cases reported Worldwide in 2011.1 Conventional first line therapy for TB comprises of a combination of isoniazid (INH), rifampicin (RIF), pyrazinamide (PZA), and ethambutol (ETB). It has been reported that out of the above mentioned drugs, INH plays major role in development of drug induced hepatotoxicity (DIH).2,3 Anti-TB DIH incidence is reported to be 9.5% in Indian patients compared to 4.3% in their western counterparts.4,5 Some of these cases can lead to severe hepatotoxicity including acute hepatic failure if not detected in time.
While exact mechanisms leading to anti-TB DIH are unknown, it has been widely suggested that polymorphisms in genes encoding certain drug metabolizing enzymes (DME) may increase the risk of DIH.6 Amongst these, N-acetyl transferase 2 (NAT2), cytochrome P 450 2E1 (CYP2E1) and glutathione S-transferase (GST) are involved in the metabolism of TB drugs.7 Apart from genetic factors, environmental factors such as advanced age, smoking, consumption of alcohol, regional food habits, socioeconomic status and malnutrition may also increase the risk of DIH.
Acetylation of INH by NAT2 enzyme, followed by oxidation by CYP2E1 enzyme leads to the generation of reactive metabolites such as hydrazine.8.–10 Recent studies suggest NAT2 and CYP2E1 genetic polymorphisms to be associated with risk of anti-TB DIH.11,12 However, few studies did not arrive at significant conclusion.13,14 It has been reported that glutathione conjugation of these species catalyzed by the GST enzyme plays a crucial role in detoxification.8,9 GST catalyzes nucleophilic attack of glutathione on electrophilic substrates thereby reducing the reactivity of potentially toxic compounds with cellular macromolecules.15,16 Of the various GST isoforms, GSTµ encoded by the gene GSTM1 located on chromosome 1p13.3 and GSTθ encoded by the gene GSTT1 located on chromosome 22q11.2 are highly polymorphic.17,18 Homozygous deletions of GSTM1 and GSTT1 are associated with depletion of glutathione content and lead to complete loss of enzymatic activity.19
The potential association of GSTM1 and GSTT1 null genotypes with anti-TB DIH has been reported in few studies’ but do not seem to arrive at a consensus. Two independent studies have reported association between the GSTM1 null genotype and DIH,20,21 while two other more recent studies rule out such association.22,23 Likewise, few studies have observed a statistically significant association between GSTT1 null genotypes and anti-TB DIH,23 while a few others do not find any association to be statistically significant.21,22,24 Therefore, the implication of GST polymorphism in anti-TB drug induced hepatotoxicity remains unclear among various populations and races.
In view of above, a case control study was designed on a large enough sample size in Western Indian population to obtain statistically significant conclusion. The objective of the study was to investigate whether GSTM1 null and GSTT1 null genotypes are associated with an increased risk of developing anti-TB DIH. Identifying a genetic marker to predict the susceptibility to anti-TB DIH can be a step towards better management and control of TB. The current study also holds significance due to challenges in treating the high incidence of tuberculosis in Indian sub-continent. There exists a genetic heterogeneity amongst various ethnic origins in Indian population.25 To the best of our knowledge’ this is the first prospective case-control study to identify GST gene polymorphism as plausible risk factor in the development of toxicity to anti-TB drugs in Western Indian population.
Material and MethodsPatient populationPatients of Western Indian cohort, newly diagnosed for active TB and undergoing anti-TB treatment were enrolled. A total of 296 patients were randomly recruited from three different hospitals: Bombay Hospital, TN Medical College and BYL Nair Hospital, and Jagjivanram Western Railway hospital located in Mumbai, Maharashtra, India. All the patients received:
- •
INH: 5 mg/kg (max. 300 mg/day).
- •
RIF: 10 mg/kg (max. 600 mg/day).
- •
PZA: 25 mg/kg (max. 2’000 mg/day).
- •
ETB: 15–25 mg/kg (max. 1,500 mg/day).
A written informed consent was obtained from each participant prior to the sample collection. The study protocol was approved by the Institutional Ethics Committee of IIT Bombay’ Mumbai, and participating hospitals and guidelines were in accordance with the Declaration of Helsinki.26
Selection criteriaNewly diagnosed tuberculosis patients from August 2010 to March 2012 were consecutively screened. These patients were evaluated in detail at baseline. Detailed history noting and clinical examinations were carried out for age, gender, height, weight, site of tuberculosis, daily intake of alcohol, drug intake, presence of symptomatic liver disease, and current symptoms or history suggestive of decompensation of liver disease. The following patients were included:
- •
Patients showing evident lesion of TB by simple X-ray or computer tomography.
- •
Positive sputum smear and /or culture for acid-fast bacilli in clinical samples.
- •
Normal alanine aminotransferase (ALT, 4-37 IU/ L), aspartate aminotransferase (AST, 4-40 IU/L) and total bilirubin levels (0.2–1.6 mg/dL).
Patients with following were excluded:
- •
Clinically and laboratory confirmed chronic liver disease such as jaundice.
- •
Positive serological testing for hepatitis B and/or C viruses.
- •
Alcoholic liver disease or habitual alcohol drinking.
- •
Patients using anti-TB drugs prior to enrollment in the study and/or other potentially hepatotoxic drugs.
- •
ALT, AST, and total bilirubin levels two fold above the upper limit of normal (ULN) before the treatment.
- •
Refusal to participate in the study.
Patients were subjected to assessment of ALT, AST, and total bilirubin levels weekly for one month and then monthly till the end of the treatment. Patients were monitored for 6–9 months according to the treatment schedule.27 According to the International consensus criteria, increase in ALT over two times of ULN or a combined increase in AST and bilirubin levels, provided one of them is above two times of ULN or any increase in ALT and/or AST above the baseline levels, was defined as anti-TB drug induced hepatotoxicity.28
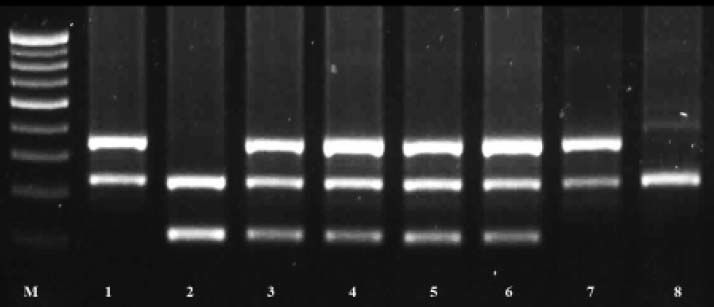
DNA isolation and genotype analysisFive milliliter of peripheral blood was collected into EDTA vacutainer (BD Biosciences, UK). DNA was isolated from whole blood using salting out method.29 Genotyping of GSTM1 and GSTT1 genes was carried out in duplicate by multiplex polymerase chain reaction (PCR) protocol using albumin gene as an internal control as described earlier with minor modification.30 Briefly, the total reaction volume was 15 µL which consists of 10X PCR buffer, 1.5 mM MgCl2, 0.5mM of dNTPs, 0.3 μΜ of each forward and reverse primer sets (Sigma Aldrich, India) for GSTM1, GSTT1 and albumin gene, 1.5U of ampli Taq Gold (ABI, Foster, CA, USA) and 100 ng of genomic DNA. PCR amplifications were performed with a Master Cycler gradient (Eppendrof, Hamburg, Germany) at annealing temperature of 60 °C. PCR products were resolved using agarose gel electrophoresis and the bands; 215, 480, and 350 bp depending on the GST genotype, were designated as GSTM1, GSTT1 and albumin, respectively. Figure 1 is a representative gel image for multiplex PCR analysis of GSTM1 and GSTT1 genotypes.
Multiplex PCR analysis of GSTM1 and T1 genes in Western Indian Population. Lane M. 100 bp DNA molecular marker (NEB, Ipswich, MA). Lanes 1 and 7. GSTM1 null genotype (215 bp). Lane 2. GSTT1 null genotype (480 bp). Lanes 3, 4, 5 and 6. Wild genotype. Lane 8. Both GSTM1 null and GSTT1null genotypes. GST: glutathione S-transferase. PCR: polymerase chain reaction.
Data for continuous variables were presented as median and interquartile range (IQR) and discrete variables as frequency and percentage. Continuous variables were compared using Mann Whitney’s U- test and discrete variables were analyzed using Fisher’s exact test. The odds ratio (ORs) and 95% confidence interval (CI) and corresponding p value was calculated using multivariate logistic regression model. The adjustments were made for potential confounders like age, gender, nutritional status [evaluated using BMI (kg/m2)], and serum transaminase level to evaluate the associations of risk factor and susceptibility. The calculations were made using R console 2.14 statistical software for windows (R Foundation for Statistical Computing, Vienna, Austria).31 All the statistical tests were based on a two tailed probability and p value < 0.05 was considered statistically significant.
ResultsEpidemiology of cases and controlsDemographic data for cases and controls are depicted in table 1. Of the 296 patients, 50 showing evidence of hepatotoxicity during anti-tubercular treatment were designated as the cases and the remaining without any evidence of hepatotoxicity as controls. All patients were HIV negative and were above 18 years of age. The percentage of females in the cases (52%) was greater than that in controls (43.5%)’ but the difference was not statistically significant. The nutritional status assessed by BMI (kg/m2) for the cases was comparable with the controls enrolled in the study.
Demographic characteristics of patients enrolled in the present study.
| Characteristics | Cases,a (n = 50) | Controls,b (n = 246) | p-value |
|---|---|---|---|
| Age (yrs) c | 37.0 (24–49) e | 36.5 (26–50) | 0.53 |
| Gender | |||
| Male d | 24 (48%) e | 139 (56.5%) | 0.28 |
| Female d | 26 (52%) e | 107 (43.5%) | |
| BMI (kg/m2) c | 21.5 (17.7–22.9) e | 21 (17.9–24.0) | 0.77 |
| Weight (kg) c | 54.0 (45–62) e | 54.5 (46–62) | 0.83 |
| Height (m) c | 1.6 (1.5–1.7) e | 1.6 (1.5–1.7) | 0.74 |
| Enzyme levels during treatment | |||
| Total bilirubin (mg/dL) c | 1.85 (0.7–5.2) | 0.9 (0.7 – 1) | < 0.001 † |
| Serum ALT (IU/L) c | 104 (74.3–182.5) | 23 (19 – 29) | < 0.001 † |
| Serum AST (IU/L) c | 124.3 (56.4–272.5) | 21 (18 – 25.5) | < 0.001 † |
Severity of hepatotoxicity was classified according to the WHO Toxicity Classification Standards.6 Accordingly’ hepatotoxicity was mild in 78%, moderate in 18% and severe in 4% of the cases with either ALT or AST levels elevated from ULN. Of the 50 cases, 36 had bilirubin levels above ULN and 12 patients amongst them showed increase in the levels of > 3 mg/dL. A statistically significant (p < 0.001) increase in total bilirubin, ALT, and AST values were observed between cases and controls during the treatment (Table 1). This corroborates the progress of cases towards toxicity due to anti-TB drugs.
Correlation of GST genotypes with anti-TB drug induced hepatotoxicityThe frequency distribution between cases and controls for GSTM1 null (*0/*0)’ GSTT1 null (*0/*0) and both GSTM1 and GSTT1 null genotypes in Western Indian population are listed in table 2. The GSTM1 null mutation was found in 21 cases compared to 61 in controls. After adjustment for potential confounding factors such as age, gender, BMI, and serum transaminase levels, a statistically significant difference in the frequency of GSTM1 null genotypes with an odds ratio of 2.14 (95% CI: 1.1–4.1, p = 0.02) was observed. However, GSTT1 null mutation was observed in 22% of cases compared to 12.2% in controls, bearing no significant association (p = 0.08). On the other hand, combination of both GSTM1 and GSTT1 null mutation was observed, 10.0 % in cases as compared to 1.63 % in controls. A statistically significant difference was observed in the frequency of both GSTM1 and GSTT1 null genotypes with odds ratio of 7.18 (95% CI: 1.7-32.6, p = 0.007).
Association of GSTM1 and T1 null genotype with the risk of anti-tuberculosis drug induced hepatotoxicity in Western Indian population.
| Genotype | Cases,a n = 50 (%) | Controls,a n = 246 (%) | ORb | 95% CI | p-value |
|---|---|---|---|---|---|
| GSTM1 | |||||
| Null | 21 (42) | 61 (24.8) | 2.14 | 1.1–4.1 | 0.02† |
| Present | 29 (58) | 185 (75.2) | |||
| GSTT1 | |||||
| Null | 1 1 (22) | 30 (12.2) | 2.03 | 0.9–4.4 | 0.08 |
| Present | 39 (78) | 216 (87.8) | |||
| Both GSTM1 and T1 null | |||||
| Null | 5 (10) | 4 (1.6) | 7.18 | 1.7–32.6 | 0.007† |
| Present | 45 (90) | 242 (98.4) |
Glutathione S-transferase (GST) is an important component of phase II drug metabolizing enzymes involved in the clearance of toxic metabolites. Of the various GST gene isoforms, GSTM1 and GSTT1 genotypes are highly polymorphic across ethnicities as well as within relatively homogenous ethnic groups.32,33 Few reports have implicated homozygous null mutation of these genes in carbamazepine and methotrexate induced liver injury.34,35
This case-control study in Western Indian population analyzed the association between GSTM1 and GSTT1 homozygous null mutations and anti-TB drug induced hepatotoxicity. The homozygous GSTM1 null genotype was significantly prevalent in cases compared to controls (42%, p = 0.02). The results were in accordance with a published report which also suggested a risk to the extent of odds ratio 2.13 (95% CI: 1.25–3.10, p < 0.05) and 2.23 (95% CI: 1.07–4.670, p < 0.03) carried out in Indian and Taiwanese population, respectively.20,21 Recent meta-analysis reports suggest that GSTM1 null genotypes to be potential risk factor for anti-TB DIH.36.–38 In contrast, to this, few other studies did not find an association between GSTM1 null genotypes and anti-TB DIH.22,39 The percentage carriers of GSTT1 null genotypes in cases were higher than control groups. We do not observe significant association between GSTT1 null genotypes and anti-TB DIH which agrees with other reported studies.21,22,24,40 However, a study carried out in Caucasians, found a significant association between GSTT1 null genotypes and anti-TB drug induced hepatotoxicity (P = 0.03).23 Putting these observations together emphasize a need for genetic association studies to recognize the predisposition in well-defined population.
Combined deletion of GSTM1 and GSTT1 gene has been reported in earlier studies.41,42 In the present study, percentage of individuals with both GSTM1 and GSTT1 null mutation were higher in cases compared to the controls (10 vs. 1.6%, p = 0.007). This shows that an individual with deletion of both copies of functional genes are at a greater risk of developing toxicity possibly due to the role played by the gene in clearance of the reactive metabolite. In contrast, earlier studies found no significant association for GSTM1 null and GSTT1 null mutation in Indian (p = 0.39) and Caucasian (p = 0.17) patients with anti-TB DIH.22,23 This may be attributed to larger sample enrolled in this case-control study compared to earlier reports.
Polymorphism in GST genes can affect the expression level of the GST enzymes43 and may increase susceptibility to certain diseases. Our findings of increased incidence of DIH in an individual having GSTM1 and double GSTM1 and GSTT1 null mutations indicate diminished enzyme activities. This may result in imperfect conjugation of reactive metabolites due to INH, partially responsible for anti-TB drug induced hepatotoxicity.20 Discrepancies in the results compared to other ethnic populations may be attributed to various factors such as different metabolism and disposition rate, ability to detoxify xenobiotics, susceptibility to certain diseases, socio economic status, and different life style habitat acquired. In addition, further study with larger population size needs to be undertaken for confirmation of our findings.
ConclusionsIn conclusion, our results suggest that GSTM1 and combined GSTM1 and GSTT1 null genotypes are significantly associated with increased susceptibility for anti-TB DIH in Western Indian population. Further, early detection of patients possessing the above genotypes may help to lower the risk of hepatotoxicity.
Abbrevations- •
95% CI: 95% confidence interval.
- •
ALT: alanine aminotransferase.
- •
Anti-TB: antitubercular.
- •
AST: aspartate aminotransferase.
- •
BMI: body mass index.
- •
CYP2E1: Cytochrome P 450 2E1 gene.
- •
DIH: drug induced hepatotoxicity.
- •
DME: drug metabolizing enzymes.
- •
ETB: ethambutol.
- •
GST: glutathione S-transferase.
- •
INH: isoniazid.
- •
IQR: interquartile range.
- •
NAT2: N- acetyl transferase 2 gene.
- •
ORs: odds ratio.
- •
PCR: polymerase chain reaction.
- •
PZA: pyrazinamide.
- •
RIF: rifampicin.
- •
TB: tuberculosis.
- •
ULN: upper limit of normal.
We gratefully acknowledge all attending physicians’ nursing staff and technical staff of Bombay Hospital, BYL Nair Hospital and TN Medical College and Jagjivanram Western Railway Hospital for assistance in collect- ing patient samples. We also gratefully acknowledge Prof. Gangamma S, NIT Surathkal for her assistance in statistical analysis.
GrantsThis work was partially sponsored by grant from Department of Science and Technology, Ministry of Science and Technology, Government of India, awarded to PPW (SR/S3/CE/11/2005).