Gallstone disease (GSD) is the result of the interaction between genetic and environmental factors and it is a major disease cause of surgery with high costs to health systems. Worldwide prevalence varies according to the ethnic population suggesting that high prevalence of GSD in certain ethnic groups is due to the presence of genetic factors implicated in different metabolic pathways. However, environmental factors play a determinant role in gene expression. This review summarizes the genes involved in biliary salt and cholesterol synthesis, lipids transport and the Lith genes. Future studies should be focused on the study of interactions between genetic and environmental factors which could be specific for each population.
Prevalence studies of GSD indicate considerable geographical and regional variations.1,2 The lowest GSD prevalence rate has been found in Asian and African populations with predominant pigment gallstones. However, the highest prevalence of GSD was found in European, Amerindian and Latin-American populations, occidentalized countries with predominant cholesterol gallstones. Pima Indians population in Chile has the most increased prevalence of GSD on 48%.1 In Mexico, necropsy of serial studies shows an approximate prevalence of 14.3% (16.2% in women and 5.6% in men).3 Also, Mexican- American women in an ultrasonographic study showed a higher prevalence of GSD on 27%.2 Therefore, prevalence variability around the world could be explain by ethnic differences, familial aggregation and the presence of a genetic background. Moreover, significant evidence of major genetic determinants of sintomatic GSD on chromosome 1p has been found in Mexican Americans.4 One study showed that genetic effects accounted for 25%, shared environmental effects for 13%, and unique environmental effects for 62%.5 Moreover, the presence of risk factors such as obesity, insulin resistance syndrome (IRS) and a family history of GSD have been associated (Table I).1,6,7-10 The aquirement of a life style similar to developed countries, characterized by a hypercaloric diet, sedentary life and stress, mixed with a particulary genetic background can determine the development of GSD. This review summarizes the genes implicated in the principal metabolic pathways involved in the development of cholesterol gallstones, such as biliary salt and cholesterol synthesis, lipids transport and the Lith genes previously described in mice.
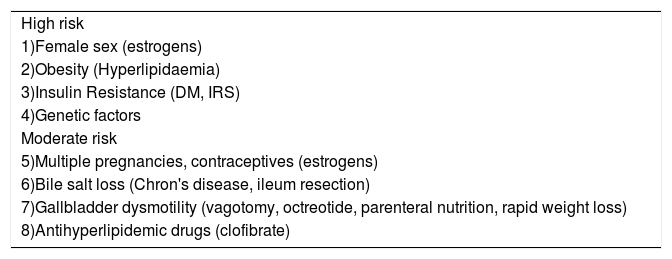
Risk factors implicated in the development of cholesterol gallstones.
| High risk |
| 1)Female sex (estrogens) |
| 2)Obesity (Hyperlipidaemia) |
| 3)Insulin Resistance (DM, IRS) |
| 4)Genetic factors |
| Moderate risk |
| 5)Multiple pregnancies, contraceptives (estrogens) |
| 6)Bile salt loss (Chron's disease, ileum resection) |
| 7)Gallbladder dysmotility (vagotomy, octreotide, parenteral nutrition, rapid weight loss) |
| 8)Antihyperlipidemic drugs (clofibrate) |
The normal bile contains 70% of biliary salts, 22% phospholipids, 4% of cholesterol, 3% of proteins and 0.3% bilirrubin. Gallstones are classified according to physical composition (Table II). Cholesterol gallstones are more prevalent in western countries, so we will be focused on the different cholesterol metabolic pathways. Hepatic cholesterol can be derived from three sources: dietary, peripheral tissues and the liver de novo synthesis. In the liver, cholesterol can be converted into primary bile acids.11,12 There are four mechanisms in the formation of cholesterol gallstones: 1) bile supersaturated with cholesterol, 2) nucleation of cholesterol monohydrate with subsequent crystal retention and stone growth, 3) abnormal gallbladder motor function with delayed emptying and stasis, and 4) gastrointestinal hypomotility modulates the enterohepatic cycling of bile salts. Precipitation of cholesterol crystals from supersaturated bile is required for gallstone formation. Therefore, the precipitation risk is directly related with the concentration of cholesterol and inversely related with the concentrations of billiary salts andphospholipids.13 The pathogenesis of supersaturated bile is related to increased hepatic cholesterol synthesis and decreased bile acid formation.14 It is also known that cholesterol contributes to the hypomotility of the gallbladder.7
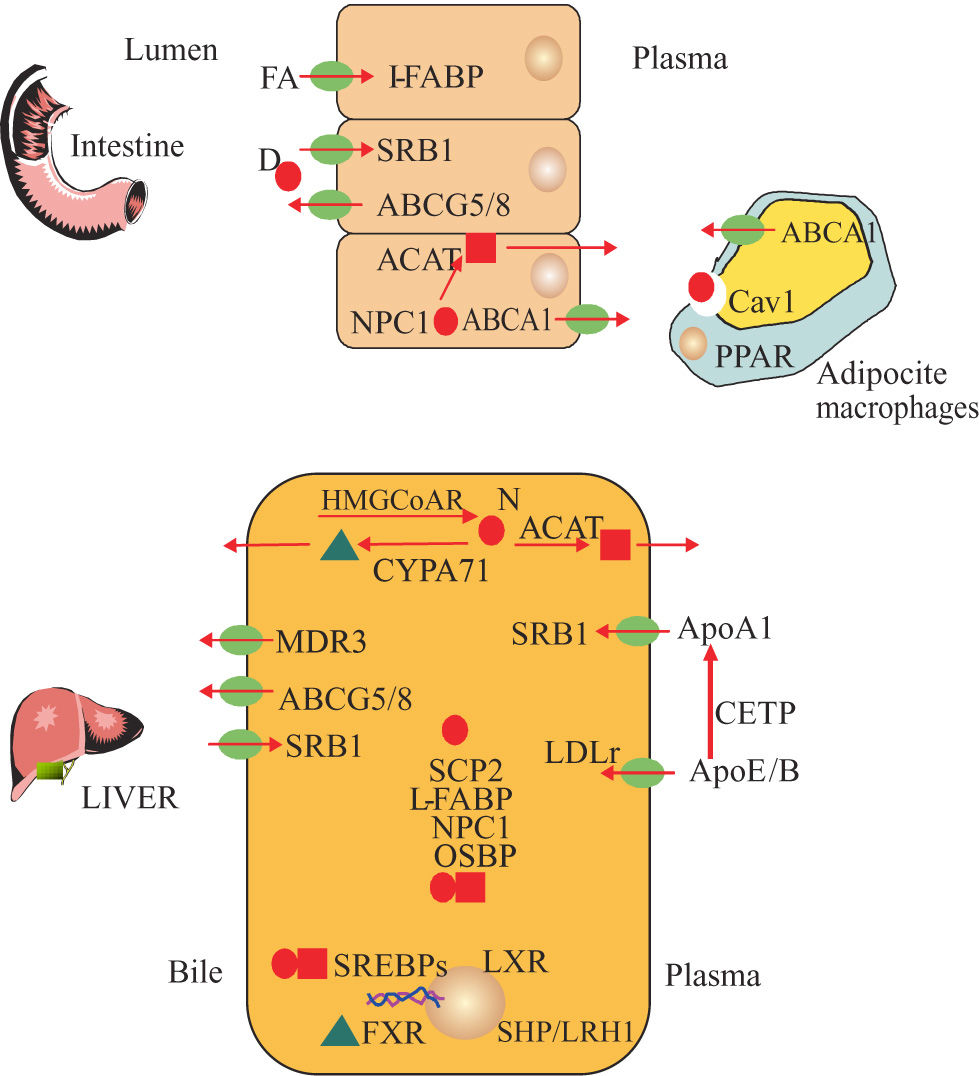
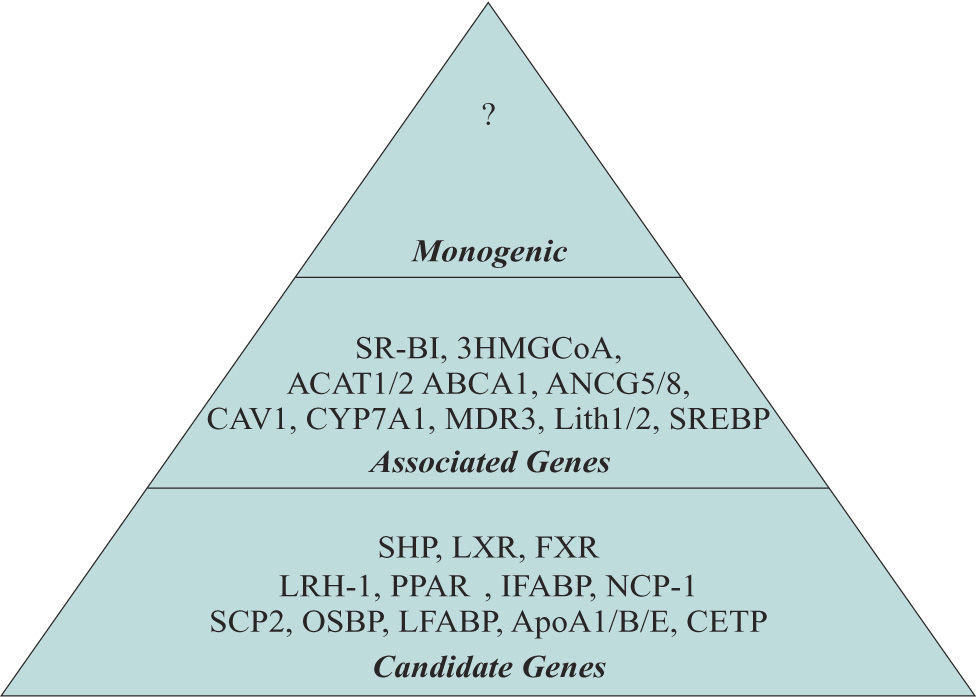
Genes involved in bile and cholesterol metabolismSeveral genes implicated in the development of GSD have been described.15 We classified them according to their participation in different pathways such as biliary and cholesterol synthesis, cholesterol transport and Lith genes identified in mice (Table III, Figures 1and2) However, other groups of genes has been identified such as Mucin genes, genes related to gallbladder functions and genes related to inflammation, possibly induced by infection with Helicobacter spp.16
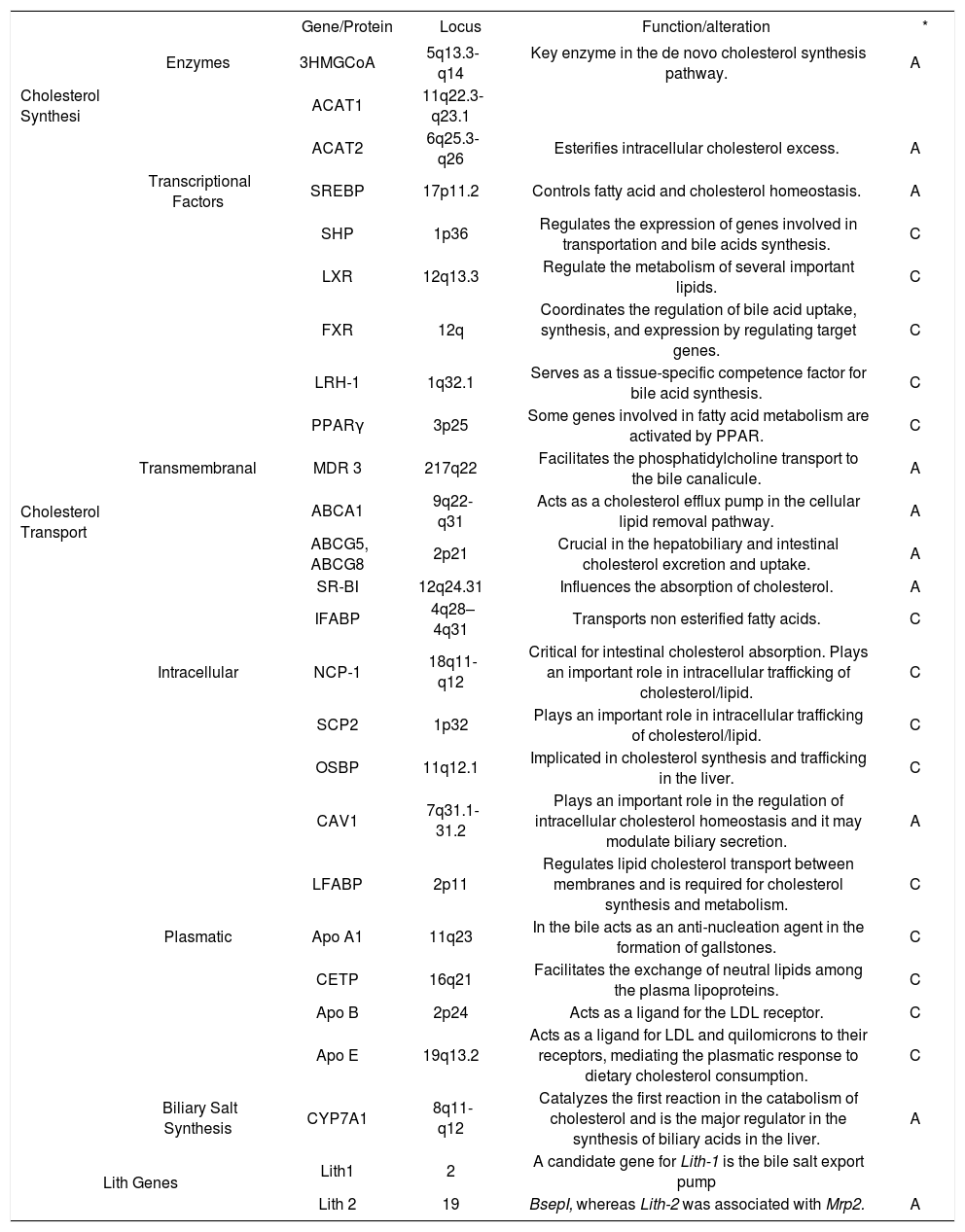
Candidate genes implicated in the development of GSD.
| Gene/Protein | Locus | Function/alteration | * | ||
|---|---|---|---|---|---|
| Cholesterol Synthesi | Enzymes | 3HMGCoA | 5q13.3-q14 | Key enzyme in the de novo cholesterol synthesis pathway. | A |
| ACAT1 | 11q22.3-q23.1 | ||||
| ACAT2 | 6q25.3-q26 | Esterifies intracellular cholesterol excess. | A | ||
| Transcriptional Factors | SREBP | 17p11.2 | Controls fatty acid and cholesterol homeostasis. | A | |
| SHP | 1p36 | Regulates the expression of genes involved in transportation and bile acids synthesis. | C | ||
| LXR | 12q13.3 | Regulate the metabolism of several important lipids. | C | ||
| FXR | 12q | Coordinates the regulation of bile acid uptake, synthesis, and expression by regulating target genes. | C | ||
| LRH-1 | 1q32.1 | Serves as a tissue-specific competence factor for bile acid synthesis. | C | ||
| PPARγ | 3p25 | Some genes involved in fatty acid metabolism are activated by PPAR. | C | ||
| Cholesterol Transport | Transmembranal | MDR 3 | 217q22 | Facilitates the phosphatidylcholine transport to the bile canalicule. | A |
| ABCA1 | 9q22-q31 | Acts as a cholesterol efflux pump in the cellular lipid removal pathway. | A | ||
| ABCG5, ABCG8 | 2p21 | Crucial in the hepatobiliary and intestinal cholesterol excretion and uptake. | A | ||
| SR-BI | 12q24.31 | Influences the absorption of cholesterol. | A | ||
| IFABP | 4q28–4q31 | Transports non esterified fatty acids. | C | ||
| Intracellular | NCP-1 | 18q11-q12 | Critical for intestinal cholesterol absorption. Plays an important role in intracellular trafficking of cholesterol/lipid. | C | |
| SCP2 | 1p32 | Plays an important role in intracellular trafficking of cholesterol/lipid. | C | ||
| OSBP | 11q12.1 | Implicated in cholesterol synthesis and trafficking in the liver. | C | ||
| CAV1 | 7q31.1-31.2 | Plays an important role in the regulation of intracellular cholesterol homeostasis and it may modulate biliary secretion. | A | ||
| LFABP | 2p11 | Regulates lipid cholesterol transport between membranes and is required for cholesterol synthesis and metabolism. | C | ||
| Plasmatic | Apo A1 | 11q23 | In the bile acts as an anti-nucleation agent in the formation of gallstones. | C | |
| CETP | 16q21 | Facilitates the exchange of neutral lipids among the plasma lipoproteins. | C | ||
| Apo B | 2p24 | Acts as a ligand for the LDL receptor. | C | ||
| Apo E | 19q13.2 | Acts as a ligand for LDL and quilomicrons to their receptors, mediating the plasmatic response to dietary cholesterol consumption. | C | ||
| Biliary Salt Synthesis | CYP7A1 | 8q11-q12 | Catalyzes the first reaction in the catabolism of cholesterol and is the major regulator in the synthesis of biliary acids in the liver. | A | |
| Lith Genes | Lith1 | 2 | A candidate gene for Lith-1 is the bile salt export pump | ||
| Lith 2 | 19 | BsepI, whereas Lith-2 was associated with Mrp2. | A | ||
Key proteins diagram of cholesterol and salt bile metabolism. Cholesterol from diet (D) and new synthesis (N)***entity***. Cholesteryl esters ***entity***, Acid Bile***entity***, Cholesterol synthesis: 3 Hidroxy Methyl Gluta- ryl CoA Reductase (HMGCoAR) Acyl Coenzyme A Cholesterol Acyl- transferase (ACAT). Regulatory Transcriptional Factors: Sterol Regulatory Element Binding Proteins (SREBPs), Liver X Receptor (LXR), Farsenoid X Receptor (FXR), Liver Receptor Homologue (LRH1), Small Heterodimeric Partner (SHP), Peroxisome Proliferator-Activa- ted Receptor Gamma (PPARy).Transmembranal transport of cholesterol and bile constituents : Multi- Drug Resistance Transporter 3 (MDR3), ATP-Binding Cassette 1 (ABCA1), ATP-Binding Cassette, Subfamily G, Member 5 and 8 (ABCG5/8), Scavenger Receptor Class B Type I (SR-BI). Intracellular trafficking of cholesterol: Niemann- Pick C1 Like 1 (NPC1L1), Sterol Carrier Protein 2 (SCP2), Oxysterol- Binding Protein 1(OSBP1), Caveolin 1 (CAV1), Liver Faty Acid Binding Protein (L-FABP). Plasmatic Transport of cholesterol: Apolipo- protein A1 (ApoA1), Cholesterol Esters Transfer Protein (CETP), Apoliprotein B (ApoB), Apolipoprotein E (ApoE) and Bile salt biosynthesis: Cholesterol 7a Hidroxilasa (CYP7A1).
The term Monogenic, Associated and Candidate genes were used to classify the genes implicated in the development of GSD. Monogenic. Already unknown and is a unique gene that promotes by his own the development of GSD. Associated genes. Participate on regulation of the principal metabolic pathways. Alteration of several of these genes could favor the development of GSD. Candidate genes. Genes indirectly involved in several metabolic pathways which could represents a risk factor for GSD.
It is the key enzyme in the de novo cholesterol synthesis pathway. Gene expression and the activity of the enzyme are regulated by intracellular cholesterol concentrations via cholesterol-derived oxysterols. In mice, cholesterol synthesis is reduced by decreasing the transcription of the genes encoding for HMG-CoAR. A relation seems to exist between HMG-CoAR expression/ activity and gallstone formation.11 However, in a human study a similar activity of HMG-CoAR was found in patients with gallstones and gallstone-free patients even though the saturation of the gallbladder bile was higher in gallstone patients.12
1.1.2 Acyl-coenzyme A Cholesterol Acyltransferase (ACAT)A cellular cholesterol sensor that esterifies the excess of intracellular cholesterol; then cholesteryl-esters are stored in cytosolic droplets or secreted into the circulation as part of lipoproteins.17 Hence, biliary cholesterol secretion is under control the activity of ACAT.2 There are two genes, the Acat1 gene encodes a thiolase mitochondrial enzyme and the cytosolic acetoacetyl-CoA thiolase encoded by the Acat2 gene. Acat genes are expressed in liver and small intestine. Acat 2 esterifies the cholesterol in murine liver and Acat 1 is the main enzyme that catalyze cholesterol-esters in humans.18,19 Hepatic Acat deficiency in rodents20 and humans22 can increase cholesterol availability for biliary secretion and the risk of GSD. Acat deficiency in the intestine diminishes cholesterol absorption resulting in the decreased biliary cholesterol output in liver and presumtive risk of GSD. Studies failed to find relationship between cholesterolosis and the amount of ACAT1 enzyme,22 suggesting the existence of others mechanisms on metabolism control of overall cholesterol.
1.2 Regulatory transcriptional factors1.2.1 Sterol Regulatory Element Binding Proteins (SREBP)SREBP1 and SREBP2 are basic helix-loop-helix leucine zipper transcription factors that regulate biosynthetic pathway of fatty acid (FA), cholesterol and LDL-R, by stimulating transcription of genes containing sterol-response-elements.23 Additionaly SREBPs regulate enzymes that generate NADPH and cytosolic acetyl-CoA, which are essential for lipogenesis.24-26 SREBP decrease transcription of the HMG-CoAR gene when high-cholesterol diet accumulates in the liver.26 Three isoforms of SREBPs have been identified. SREBP1a and SREBP1c are transcribed from a single gene; SREBP2 is transcribed from a second gene. In liver and other organs, SREBP1c and SREBP2 are most highly expressed. SREBP1 is synthesized as a precursor; in sterol-depleted cells, the precursor is cleaved to generate a soluble fragment that translocates to the nucleus.27 Sterols inhibit the cleavage of SREBP1. Proteolytic release requires a sterol-sensing protein (SCAP).28
1.2.2 Small Heterodimeric Partner (SHP)SHP heterodimerizes with several nuclear receptors and it is expressed in the liver and intestine. It has been demonstrated that activation of FXR (see ahead), by natural and synthetic agonists, increases SHP levels, which in turn reduces SREBP-1c expression29-31 mechanism proposed to explain the reduction of CYP7A1 expression by bile acids, which also invoked SHP as a mediator.32 SHP is also a potent repressor of LRH1 and its target genes. Together, SHP and LRH1 are important factors in the regulation of gene expression involved in transport and synthesis of bile acids, thereby influencing the formation of supersaturated cholesterol.29
1.2.3 Liver X Receptor (LXR)Two members are known in this subfamily: LXR (NR1H3) and LXR (NR1H2). Both have a different expression pattern and form heterodimers with another nuclear receptor, the retinoic X receptor (RXR). Whereas LXR (NR1H3) is ubiquitously expressed, LXR (NR1H2) is most highly expressed in the liver and intestinal tract which are the tissues most involved in cholesterol metabolism.30 The expression of the LXR target genes are lowered via SHP-dependent mechanism.32
1.2.4 Farsenoid X Receptor (FXR)FXR forms part of the RXR that binds with high affinity to bile acid.33 Bile acids are able to activate FXR which regulates the target genes in bile acid uptake, synthesis, transport and cholesterol metabolism. Identified target genes are CYP7A1.34,35 ApoC-II,36 CYP8B1, basolateral sodium taurocholate cotransporter protein, hepatic canalicular bile salt transporter, ileal bile acid binding protein, SHP and phospholipid transfer protein.29,30 There is evidence that the production of certain apolipoproteins is regulated by the bile acid–activated nuclear receptor FXR.36,37
1.2.5 Liver Receptor Homologue (LRH1)Also known as NR5A2, it belongs to the NR5A or the Ftz-F1 subfamily of nuclear receptors and is expressed in liver, pancreas, intestine, and ovary.38 LHR-1 is an orphan nuclear receptor that binds as a monomer. LRH1 serves as a tissue-specific competence factor for bile acid synthesis. LRH1 target genes include: α-fetoprotein, SHP, CETP, CYP7A1 and CYP8B1.29
1.2.6 Peroxisome Proliferator-Activated Receptors(PPARs)Is a nuclear receptor activated by hypolipidemic compounds like fibrates, but FA have been found to be the natural ligands. Genes involved in FA metabolism and β-oxidation have been found to be activated by PPAR. Specific PPARγ target gene is peroxisomal acyl-CoA oxidase.12 PPARγ and molecules like PPARγ coactivator-1 are involved in inflammatory gallbladder39 and cholesterol formation.40
2. Cholesterol transport2.1 Transmembranal transport of cholesterol2.1.1 Multidrug Resistance Transporter 3 (MDR3)MDRs are P-glycoproteins and were discovered as large cell membrane proteins overproduced in cancer cells resistant to a diverse set of hydrophobic drugs.41 MDR3 gene is separated from the MDR1 gene by 34 kb. Both human genes are transcribed in the same direction, MDR3 being located downstream from MDR1.42 MDR3 codes for a phosphatidylcholine transporter protein in the membrane of the hepatocyte that facilitates the phosphatidylcholine transport to the bile canalicule. Certain authors have reported cases of acute pancreatitis caused by microlithiasis due to mutations in the MDR3 gene, resulting in a phosphatidylcholine deficiency associated with a rapid crystallization of cholesterol.43
2.1.2 ATP-Binding Cassette 1 (ABCA1)Is a cholesterol and phospholipid efflux pump mediated by ApoA1, essential for HDL formation and controls the first step of reverse cholesterol transport.14,44 Overexpression in transgenic Abca1 mice increases plasma HDL-cholesterol levels, hepatic delivery of HDL cholesterylesters and biliary cholesterol concentrations.45 Tangier disease is caused by mutations in the ABCA1; these patients have a massive tissue deposition of sterols with near to zero plasma levels of HDL.46 In gallbladder epithelial cells, ABCA147 is regulated by LXR and RXR48 and modulates biliary cholesterol concentrations and its excretion from the body.14 Retinoids and others ABCA1 regulators offer a novel class of agents for treating elevated cholesterol or prevention of GSD in rodents. Aramchol is a FA-bile acid conjugate that induces ABCA1-dependent cholesterol efflux without affecting transcriptional control.49
2.1.3 ATP-Binding Cassette, Subfamily G, Member 5 and 8 (ABCG5/8)The second set of ABC-half transporters implicated to have a role in the physiological pathways by which dietary cholesterol, as well as non-cholesterol sterols, traffics in human body.50,51 The two genes are tandemly grouped.52 The ABCG5/8 couple is crucial for hepatobiliary and intestinal cholesterol excretion, expressed in enterocytes and the canalicular membrane.51,53 Liver and intestine maintain sterol balance with respect to noncholesterol sterols.54 In the liver, they are the main player in the secretion of cholesterol and sterols into the bile.55 Mutations of these genes increases dramatically the plasma and hepatic cholesterol levels in response to changes in dietary cholesterol content and cause a rare human disorder, sitosterolemia.13,52,56 This disorder has been identified in Mexican-American patients with high concentrations of sterols in plasma tissues52 indicating high risk for GSD.57
2.1.4 Scavenger Receptor Class B Type I (SR-BI)This protein is a multifunctional receptor able to bind to anionic phospholipids, native and oxidized LDL and apoptotic cells. It has affinity for HDLs and mediates the selective uptake of cholesterol-esters. It is expressed in intestine and it has been suggested that it contributes to the entrance of cholesterol in the body.58 Polymorphisms studied are associated with variation in plasma concentrations of fasting triglyceride.58 It has been suggested a possible mechanism involved in the absorption of cholesterol in gallbladder synergized by the union of ApoA1 also present in the bile.59
2.1.5 Intestinal Fatty Acid Binding Protein (IFABP)The FABP2 gene belongs to a family expressed in a tissue-specific manner.60 Protein IFABP is expressed only in epithelial cells of the small intestine,61 transporting non esterified FA from the plasma membrane, through the aqueous cytosol, to the endoplasmic reticulum (ER),62 influencing lipid absorption and plasma levels of lipids.63-65 FABPs are also hypothesized to serve as cytosolic FA carriers to transport FAs among cellular organelles where FAs have various functions. As intracellular transporters, FABPs deliver regulatory lipids to the nucleus of the cell where the lipids can influence PPAR mediated gene expression.66
2.2 Intracelullar transport of cholesterol2.2.1 Niemann Pick Type C-1 (NCP1)This gene codes for transmembranal protein Niemann- Pick C1 Like 1 (NPC1L1), localized in jejunal enterocytes that is critical for intestinal phytosterols and cholesterol absorption containing a sterol sensing domain homologous to the domains found in HMGCoAR.67 NPC1L1 is also a peripheral cholesterol transporter for the energy-dependent vesicular trafficking process of endocytosed lipoprotein cholesterol.68-70 Hepatic NPC1L1 is an important factor that regulates biliary cholesterol secretion in mice, because its inactivation produces an impaired biliary cholesterol secretion in cholesterol-fed mice.71
2.2.2 Sterol Carrier Protein 2 (SCP2)This protein plays an important role in intracellular trafficking of cholesterol/lipids72-74 specially to mitochondria.75 Different authors localize the SCP2 in peroxisomes and others in the cytosol.76 SCP2 contains both a thiolase domain and a sterol carrier-protein domain and is the key enzyme in β-oxidation of bile acid intermediates. SCP2 is necessary for the rapid transport of newly synthesized cholesterol into bile as well as the conversion of free cholesterol into cholesterol-esters.76,77 Increased hepatic SCP2 expression correlated with biliary cholesterol hypersecretion78,79 in human patients80 and development of GSD in mice.81
2.2.3 Oxysterol-Binding Protein (OSBP)A citosolyc protein involve in cholesterol synthesis and trafficking in the liver. OSBP transports sterols from lysosomes to the nucleus, where sterol downregulates genes like LDL-R, HMG-CoAR, HMGCoAS. OSBP regulates cellular transport of cholesterol, sphingomielyn, oxysterol and sterol by secretory vesicles and control of signalling cascades. OSBP acts as a cholesterol-binding scaffolding protein coordinating the activity of phosphatases to control the extracellular signal-regulated kinase-signaling pathway.82-84
2.2.4 Caveolin (CAV 1)CAV1 is highly expressed in intrahepatic basolateral and canalicular membranes. CAV1 mediates endocytosis by caveolae, which are plasma membrane invaginations that are highly enriched in cholesterol and sphingomyelin.85,86 CAV1 is implicated in cell signaling, transcytosis and in regulation of intracellular cholesterol transport.87 In hepatocytes, the SRBI has been shown to be associated with CAV1 indicating their role in cholesterol uptake. Lipid droplets are potential target organelles for caveolar endocytosis demonstrating a role for CAV1 in the maintenance of free cholesterol levels in adipocytes.88 The transcription of Cav1 is under the positive control of SREBP pathway, increasing when intracellular cholesterol levels are high.89 PPARγ overexpression cans upregulate Cav1 expression in macrophages. CAV1 facilitates the transport of cholesterol from the ER to the plasma membrane and it may modulate biliary secretion.90
2.2.5 Liver Fatty Acid Binding Protein (L-FABP)L-FABP is an abundant cytoplasm component of the hepatocyte that regulates lipid cholesterol transport between membranes. It is required for cholesterol synthesis and metabolism13,91 and responsible for the diffusional mechanism of FA transfer to membranes.92 L-FABP expression is regulated by PPARγ.93 Polymorphism T94/T94 exhibit higher ApoB levels whereas carriers of the A94 allele seem to be protected against high ApoB levels when consuming a saturated fat diet.94
2.3 Plasmatic transport of cholesterol2.3.1 Apolipoprotein AI (ApoAI)Apo AI is the main apolipoprotein on HDL. ApoAI is a cofactor for Lecitin Cholesterol Acyl-Transferase (LCAT), which is responsible for the formation of most cholesteryl-esters in plasma. ApoAI is related to CAV1 and both are involved in the regulation of intracellular cholesterol trafficking for the assembly of cellular lipids to ApoAI-HDL.95 ApoAI promotes efflux of cholesterol from cells. ApoA1 knockout mice have low plasma HDL-cholesterol levels and their rate of hepatic cholesterol synthesis is 50% lower than wild-type mice. ApoA1 and ApoA2 are secreted in to bile,96 and bile acids influence expression of ApoAI.97 In contrast, ApoA1 overexpressing mice have been reported to have a 2-fold increase in biliary output of bile acid and cholesterol. In humans, it has been observed that ApoAI removes certain lipids from the bile and acts as an anti-nucleation agent in the formation of gallstones.98
2.3.2 Cholesterol Esters Transfer Protein (CETP)This protein facilitates the exchange of neutral lipids among the plasma lipoproteins and induces a transfer of cholesterol-esters of the HDL toward the lipoproteins rich in tryglicerydes (TG) in exchange for TG. An enzimatic deficiency causes hyperalphalipoproteinemia and the G to A sustitution in the intron 14 splice donor is a common mutation. Therefore, a high activity of CETP could decrease levels of HDL-cholesterol and high levels of TG, a lipid pattern that increases the risk to develop GSD.99
2.3.3 Apolipoprotein B (ApoB)ApoB is the main protein of chylomicrons and LDL. There are two main forms: ApoB48 and ApoB100. The first is synthesized exclusively by the gut and the second by the liver. ApoB acts as a ligand for the LDL-R mediated by endocytosis. ApoB has been associated with familial hypobetalipoproteinemia, an autosomal dominant disorder of lipid metabolism characterized by extremely low plasma levels of ApoB, as well as low levels of LDL and total cholesterol.100 The XbaI polymorphism is associated with differences in plasma LDL-cholesterol levels and contributes relatively in the development of GSD in certain populations.101
2.3.4 Apolipoprotein E (ApoE)It is a major protein component of VLDL and minor of HDL. ApoE acts as a ligand for LDL and quilomicrons to their receptors mediating the plasmatic response to the dietary cholesterol. In familial type III hyperlipoproteinemia, there is impaired clearance of chylomicron remnants and VLDL, increased plasma cholesterol and TG due to a defect in ApoE.102 ApoE has three allelic variants: E2, E3 and E4. Carriers of E2 allele present low concentrations of total cholesterol and LDL-cholesterol in plasma, while E4 allele carriers present higher levels of LDL and total cholesterol that causes differences in the affinity to ligand-receptor binding. Carriers of E4 allele have high risk of GSD because it increases the lipoprotein uptake and consequently the hepatic and biliary concentration of cholesterol.103,104 In contrast, E2 allele provides a protection against GSD.98
3. Bile salt biosynthesis3.1 Cholesterol 7α Hidroxilasa (CYP7A1)The first reaction in the catabolism of cholesterol is mediated by the enzyme CYP7A1 that produces biliary acids. LRH1 and RXR/FXR heterodimers regulate CYP7A1 expression and bile acid synthesis.48 A deletion in these gene results in the loss of the function of the enzyme increasing the levels of LDL that predispose to premature GSD due to the inability for cholesterol solubilization in the biliary salts. Several studies suggest that a substitution of A for C in the position -204 of the promoter of the gene CYP7A1 has been associated with variations in the concentrations of LDL-cholesterol. In men, the C variant has been associated with an increased rate of total cholesterol/HDL index and higher levels of LDL-cholesterol in plasma.105
4. Lith genesGenetic era in GSD research began with the detection of the first cholesterol gallstone genes (Lith) by quantitative trait Loci (QTL) mapping in crosses between gallstone-susceptible (C57L/J) and gallstone-resistant (AKR) inbred strains of mice.106 QTL analysis localizes additional unknown gallstone genes. It provides the genetic basis for the orthologous human. Lith genes might encode lipid transporters in the canalicular membrane that could transfer lipid molecules into hepatic bile or regulators of cholesterol metabolism. The major murine cholesterol gallstone QTLs determined are Lith1 (chromosome 2), Lith2 (chromosome 19), Lith3 (chromosome 17), Lith4 (chromosome X) and Lith5 (chromosome 5; Muc3).21 The most important Lith genes associated with GSD are Lith-1 and Lith-2. These inbred mice present alterations in some of the cholesterol regulatory genes mentioned above as SRBI, ACAT2, HMG-CoAR, CYP7A1(1) and Hepatic Lipase.107
ConclusionsGenetic factors identified in animal models suggest plays an important role accounting 25% in the development of GSD.4 This review summarizes the most important genes implicated in such pathology. We conclude that it is necessary to study the polymorphic genes (for example: 3HMGCoA, CYP7A1, CETP, Apo A1/B/E, ABCG5/ 8) due to variability and higher prevalence present in each population. However, the presence of environmental factors (diet, physical activity and emotions) of each culture could determine gene expression and exerts an independent effect in the development of GSD. Therefore, integral studies about the interaction between specific genetic and environmental factors in each population will be a very important approach to develop new strategies for prevention, diagnosis and management of patients with GSD based on personalized medicine (genomic medicine).