Chronic liver diseases account for a considerable toll of incapacities, suffering, deaths, and resources of the nation's health systems. They can be prevented, treated or even cured when the diagnosis is made on time. Traditional liver biopsy remains the gold standard to diagnose liver diseases, but it has several limitations. Liquid biopsy is emerging as a superior alternative to surgical biopsy given that it surpasses the limitations: it is more convenient, readily and repeatedly accessible, safe, cheap, and provides a more detailed molecular and cellular representation of the individual patient's disease. Progress in understanding the molecular and cellular bases of diseased tissues and organs that normally release cells and cellular components into the bloodstream is catapulting liquid biopsy as a source of biomarkers for diagnosis, prognosis, and prediction of therapeutic response, thus supporting the realization of the promises of precision medicine. The review aims to summarize the evidence of the usefulness of liquid biopsy in liver diseases, including the presence of different biomarkers as circulating epithelial cells, cell-free nucleic acids, specific species of DNA and RNA, and the content of extracellular vesicles.
Chronic liver diseases (CLD) is a major global public health problem estimated to affect 844 million people and cause 2 million deaths per year [1]. In contrast to many chronic diseases, a large proportion of CLD can be prevented, treated and/or even cured if diagnosed early which makes methods for timely diagnosis critically important [2]. Among the CLD, liver cancer is the second leading cause of cancer-related death globally [3]. Among all primary liver cancers, hepatocellular carcinoma (HCC) is the most common, with approximately 90% of cases [4]. Traditional liver biopsy remains the gold standard to diagnose CLD and assess the pattern and severity of disease in individual patients, because currently available non-invasive tests, including imaging and serological methods, lack the sensitivity and specificity to identify early stages of fibrosis and are not useful in the discrimination of inflammation from hepatocellular injury [5]. While tissue biopsy is an important and useful diagnostic tool, it is both invasive and demanding of significant sample preparation and evaluation by a pathologist, variations in both sampling and interpretation, plus the limitation of the analysis to the disease stage when performed do not provide the level of detailed molecular information-theoretically possible about an individual patient's disease.
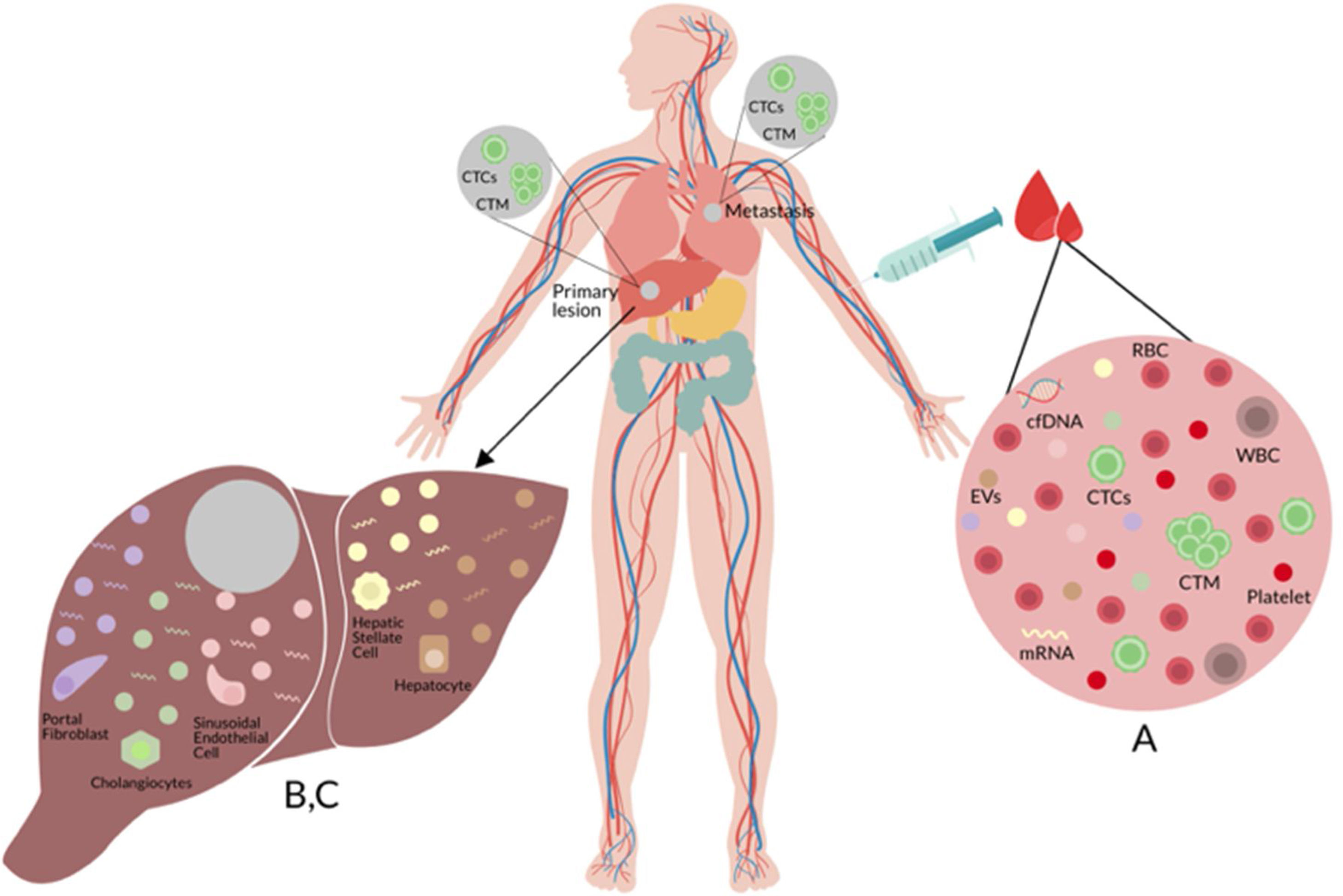
In light of these limitations on the use of single biopsies, new ways to observe tumor genetics and tumor dynamics have evolved. In 1948, the publication of a manuscript that described circulating free DNA (cfDNA) and RNA in the blood of humans was, without knowing it, the first step toward the ‘liquid biopsy’ [6]. Liquid biopsy has its origins and major progress in oncology. The National Cancer Institute defines liquid biopsy as a test done on a sample of blood to look for cancer cells from a tumor that are circulating in the blood or for pieces of DNA from tumor cells that are in the blood [7]. Numerous studies have demonstrated the feasibility of analyzing material from tumor cells circulating in the bloodstream, which could provide readily accessible, accurate, and dynamic information to diagnose disease and evaluate its progression [8]. In recent years, the results of the Human Genome Project and pharmacogenomics research overcame the old paradigm of ‘one size fits all’, providing a large amount of molecular data that generated the concept of “precision medicine” with the aim of tailoring therapies for patients according to their specific pathology, based on the development of biomarkers; and liquid biopsy has been proclaimed with the ability of detect biomarkers that carry information about the tumor advancement [9] (see Fig. 1).
(A) Liquid biopsy of hepatocellular carcinoma (HCC): Spontaneous circulation of CTCs and CTM in peripheral blood reflects tumor progression and tumor spread in patients with HCC [20]. (B) Cell-free nucleic acids (cfNA): cfNA (DNA and RNA) are known to come from apoptotic and necrotic cells or to be released from living eukaryotic cells, thereby providing a valuable source of material which can instruct about natural or biological and pathological processes within its cellular source [170]. (C) Extracellular vesicles (EVs): EVs are small membrane vesicles released by cells in the extracellular environment as part of normal physiology or during pathological processes, which function is the communication between cells, their cargo (mRNAs, miRNAs, proteins and lipids) may reflect the cell of origin as well as the specific stress that induces their formation and release [149,150]. Red blood cell (RBC), white blood cell (WBC), circulating tumor cells (CTCs), circulating tumor microemboli (CTM), and cell-free DNA (cfDNA).
In contrast with surgical or solid biopsy, liquid biopsy has considerable advantages; for example, it is a minimally invasive procedure that can be performed routinely without extensive training, allows repeated sampling that can offer disease monitoring over time, and the molecular analyses of nucleic acids in samples capture better the overall genetic complexity of lesions. Besides, samples are easier to process and store and are cheaper than the classic biopsy procedures and may be more effective for early disease detection due to the sensitivity and selectivity of current molecular methodologies [10].
This review provides a summary of the present status and future potential for the use of liquid biopsy as a diagnostic, predictive and prognostic tool of particular value in the prevention of CLD.
2Circulating epithelial cells2.1OverviewLiquid biopsy can detect circulating epithelial cells (CECs) in the setting of localized cancers as well as preneoplastic lesions, suggesting their presence is not limited to cells derived from established cancers [11,12]. Hepatic CECs have not been widely studied in the absence of malignancy, but existing studies suggest they could serve as a powerful biomarker for the widespread diagnosis and monitoring of various CLDs as well of hepatocellular carcinoma. The CECs associated with a known malignancy are termed circulating tumor cells (CTCs) [13] (see Fig. 1).
CTCs are lost by both primary and metastatic cancers and they are thought to mediate the hematogenous disperse of cancer to remote sites, including bone, lung, brain, and liver. In metastasis, whole living cells (CTCs) released from carcinoma in situ survive in the bloodstream to then successfully settle in another organ [14]. The pathophysiology of metastatic cell distribution, most likely, involves interaction with other circulating cells and its target site.
Despite considerable progress, studies of CTCs and their potential use for diagnosis and monitoring of disease, some difficulties remain. One of the main issues in oncology is that in early-stage cancer, few tumor cells are present in the bloodstream because their number is proportional to tumor mass [15]. The estimated abundance of CTCs is almost to none in metastatic disease, e.g., a blood sample from these patients may only contain 1–10 cancer cells in a sea of blood cells [16]. The sensitivity and specificity of the detection techniques for CTCs, which have not yet been developed, must be standardized to obtain reliable and reproducible findings. Isolation of pure populations of CTCs is needed for accurate molecular characterization needed for useful clinical evaluation of CTCs. The technologies required for CTC detection pivot on a large volume of blood and this impedes clinical use in daily practice [17].
However, tumor-derived CTCs in the bloodstream are a heterogeneous population, and therefore, more likely than a tissue sample to reflect the composition of the whole tumor in terms of phenotypic and genetic makeup [18,19].
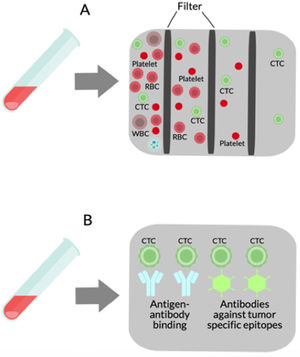
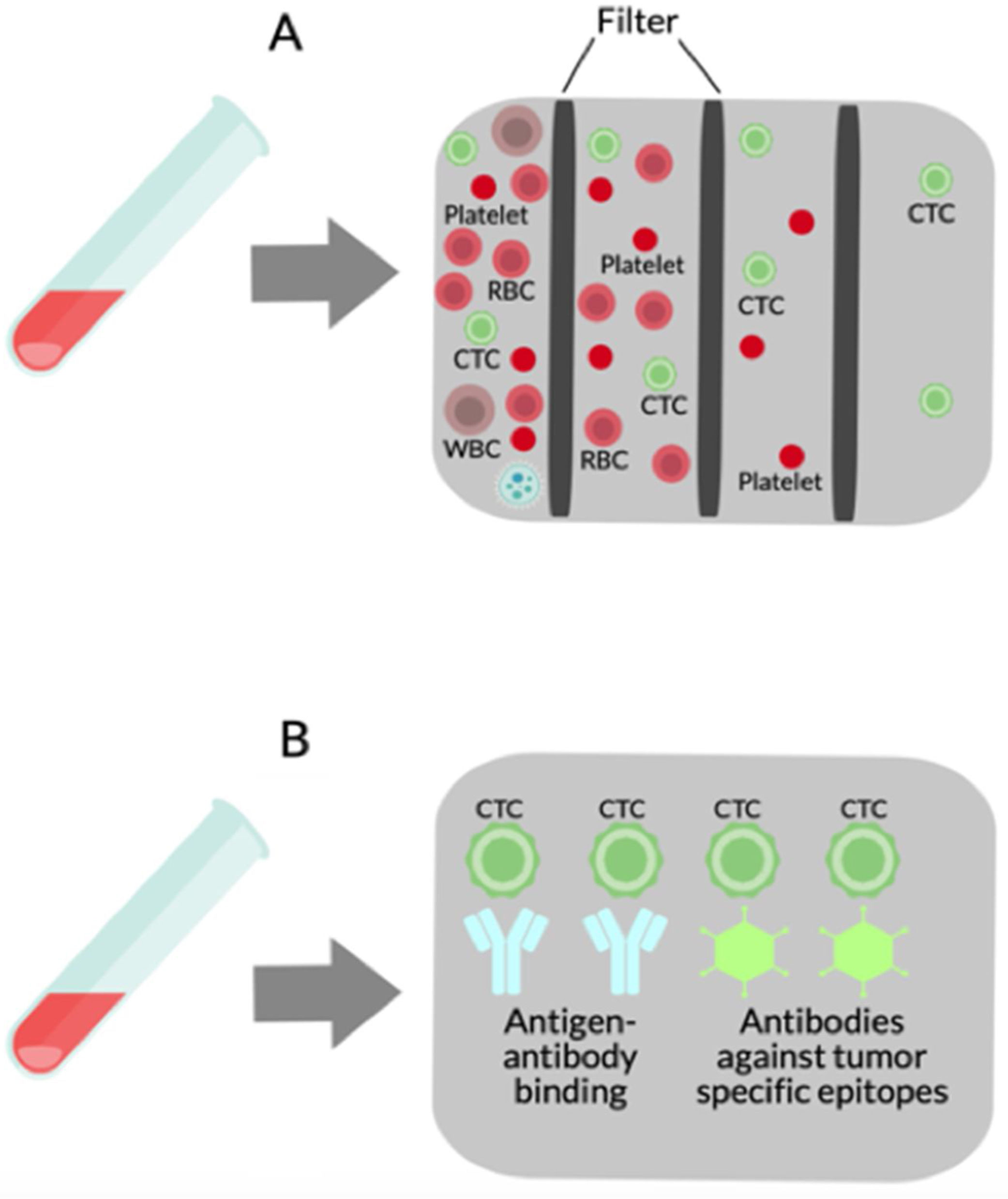
2.2Isolation of CTCsThe methodology used to isolate or detect CTCs is divided into physical and biological procedures (see Fig. 2).
Isolation methods for CTCs. (A) Physical methods use multiple filters based on size, density, migratory capacity, deformability, electric charge, and polarity. (B) Biological methods depend on antigen–antibody binding and antibodies against tumor-specific epitopes including epithelial cell adhesion molecule (EpCAM), human epidermal growth factor receptor 2 (Her2), and prostate-specific antigen (PSA) are typically used in CTCs purification.
Physical procedures rely primarily on the CTCs physical properties, such as size, density, migratory capacity, deformability, and electric charge; newer methods are also based on polarity, which has recently been shown to be critical for adhesion, attachment, migration, and metastasis of CTCs [20,21]. These methods employ centrifugation and filter or flow devices with channels of different sizes and/or properties and can be used to separate and isolate both normal and cancer cells [22]. Most CTCs originate from epithelial tumors and must be separated from much greater numbers of cells in the mix of blood samples, and for that reason, several filtration-based techniques have been developed [23,24] since this methodology allows to process large volumes of blood. There are substantial differences in cell size among cancer patients [25], new techniques with several filters were developed to isolate a wide range of CTCs sizes [26]. Vona et al. first used the isolation by size of epithelial tumor cells (ISET) method to detect CTCs in HCC patients. ISET is a low-cost method that involves the binding of CTCs to membranes and does not require special equipment. However, a potential problem with ISET devices may be a difficulty to completely release bound CTCs from the membranes used in the isolation process, which may yield invalid genetic analyses of the cell populations prepared by this method [22]. Some devices isolate cancer cells based on their size and deformability. For example, Mohamed et al. made an apparatus with narrow channels that filter only cancer cells from other blood cells circulating [27].
Using physical isolation methods followed by cytomorphologic analysis it was shown that the patterns of spontaneous circulating CTCs and circulating tumor microemboli (CTM) in peripheral blood reflect tumor development and metastasis in HCC patients. Also, the numbers of CTCs and CTM were associated with survival in time in HCC patients [20].
Biological methods for CTC isolation depend on antigen-antibody union using antibodies against tumor-specific epitopes. Examples that have been used for CTC purification include epithelial cell adhesion molecule (EpCAM), prostate-specific antigen (PSA), and human epidermal growth factor receptor 2 (Her2) [28]. EpCAM is the most generally used in CTCs purification because its expression is global in cells of epithelial origin, but it is missing in other blood cells. The US Food and Drug Administration (FDA) has only approved EpCAM analysis for use in patients with prostate, breast and colorectal cancer. Cell-Search system (Veridex LLC, NJ, USA) is the most commonly used CTC platform for EpCAM analysis. In this platform, EpCAM antibodies coated with immunomagnetic beads are used to seize CTCs that are then immunostained with two positive markers (cytokeratins 8/18/19 and 4′6′-diaminidino-2-phenylindolehydrochloride) and a negative marker (CD45) [29–34]. Using the Cell-Search system, Sun et al., and Guo et al., proposed EpCAM+CTCs in early decision-making treatment, monitoring treatment response, and to indicate in HCC recurrence [35,36]. Using antibody-based procedures Sun et al. compared CTCs isolated from peripheral veins, portal vein, and hepatic vein, before HCC resection. The greatest number of CTCs was detected in the hepatic vein exiting the liver, with a dramatic reduction in peripheral vessels after passage through the lungs. Furthermore, characterization of CTCs confirmed phenotypic heterogeneity across vascular sites, with a predominantly epithelial phenotype in the hepatic vein, versus an epithelial-mesenchymal transition (EMT) type (associated with Smad2 and beta-catenin) of CTCs isolated from the peripheral veins [37]. The liquid biopsy also offers the opportunity to characterize CTC interactions with circulating immune cells, and the combination of elevated EpCAM-positive CTCs and an increased number of regulatory CD4+ T cells have been reported to be indicative of early HCC recurrence and poor clinical outcome [38].
An alternative method for CTC isolation that offers high sensitivity and specificity is a microfluidic-based CTC-chip platform. In this platform, CTCs may be isolated accurately from limited volumes of blood by flow through microposts coated with EpCAM antibodies. The advantage of this method is that it enhances the interactions between CTCs and the antibody-coated surface and dynamic flows to ban nonspecific binding [39,11]. More alternatives implemented are the CTC-chip to improve the CTCs purification followed by an off-chip enzymatic treatment to enhance the pureness rate, along with a ligand-receptor binding assay to increase the capture rate [40,41]. Also, Wang et al. suggested that the CTC-chip platform might be a convenient method for the efficient and simple detection of CTCs in HCC patients. They created a CTC-BioT-Chip, consisting of a biocompatible and transparent HA/CTS (hydroxyapatite/chitosan) nanofilm coated by sLex-AP (aptamer for carbohydrate sialyl Lewis X) and demonstrated that it could be useful to predict patients prognosis [42]. Most recently, Oklu et al. introduce an antigen-agnostic cell sorting instrument called the iChip, which confines CECs protecting high-quality RNA content and cell viability. Liver-specific RNA markers combined with the iChip made possible a two-step assay for the enrichment and detection of CECs in HCC and CLD [43].
The most relevant clinical applications that have been demonstrated, until now, are presented below. Matsumura et al., using real-time polymerase chain reaction (RT-PCR) demonstrated that alfa fetoprotein (AFP) mRNA in CTCs could be a predictor of outcome in HCC patients [44]. Mou et al. predict the relapse and prognosis of HCC patients using melanoma-associated antigens, MAGE-1 and MAGE-3 mRNAs, as markers of CTCs in the blood [45]. Yao et al. demonstrated that the overexpression of glypican protein-3 (GPC-3) mRNA is useful as a clinical biomarker from early detection and evaluating metastasis on HCC [46]. CD44 and K19 mRNAs have been shown as cancer stem-cell markers in HCC, used as a prognostic factor demonstrated by Choi et al. [47]. Liu et al., and Bahnassy et al., established that intracellular adhesion molecule 1 (ICAM-1), cytokeratin 19, CD133, CD90, and telomerase in HCC blood samples have prognostic value in HCC [48,49]. In the case of benign diseases, Kelley et al., described the detection of CTCs’ DNA in patients with HCC, to a level of at least two cells per 7.5ml, but not in patients with CLD without HCC [50].
2.3CTCs escape from immunosurveillanceCTCs may have a persistent interaction with blood cells that enables them to escape immunosurveillance. Even though certain blood cells are thought to be a key factor in cancer defense, their interactions with CTCs may be a mechanism to escape immune surveillance at times. For example, interactions with T cells can protect CTCs and promote their survival [19]. This can occur when β1-integrins are expressed on CTC surfaces; this facilitates CTC capture by neutrophil extracellular traps (NETs) and has the effect of developing a metastatic niche [51]. Another mechanism by which CTCs escape immune surveillance is their binding to receptors glycoprotein Ib (GPIb) on platelets which promotes tumor cell extravasation that results in metastatic colonization [52].
3Cell-free DNACell-free DNA (cfDNA) detected in blood offers a less invasive and a replicable liquid biopsy, giving real-time dynamic analysis and evaluation of disease emergence, evolution, and therapy assimilation with a huge impact in the medical field [53]. The cfDNA released by a tumor is called circulating free tumor DNA (ctDNA), although, in patients with cancer, cfDNA reflects a mixture of wild-type nonmalignant cfDNA and ctDNA [54]. The presence of ctDNA in the bloodstream may provide two types of information. Quantitative changes reflect disease activity, response to treatment and even recurrence, while qualitative changes on genomic abnormalities (DNA copy number variations, gene mutations, tumor-specific methylation patterns, loss of heterozygosity, and/or microsatellite instability) may reflect tumor evolution during disease progression [55–58].
In the case of quantitative analysis, Huang et al., and Chen et al., reveal that cfDNA quantity was extremely higher in HCC patients and linked with a bad prognosis; however, the high levels of cfDNA were not specifically derived from HCC which may limit the use of quantitative measurements of cfDNA [59,60]. Nevertheless, Chen et al. found that HCC patients had significantly elevated levels of cfDNA in their serum, however, the sensitivity was low [60]. In HCC Patients with Hepatitis C Virus (HCV), Tokuhisa et al. noted cfDNA to be on the rise. They also found that high cfDNA serum levels post-HCC resection were an independent predictor of shorter overall survival and the presence of distant metastases after hepatectomy [61]. In another study of the same group, cfDNA levels correlated with aspartate aminotransferase levels, inflammatory cytokine gene expression, and neutrophil levels, hinting a correlation of inflammation degree in the primary tumor and the cfDNA levels [62]. Iizuka et al., and Ren et al., reported a significant association between ctDNA serum levels, tumor size, TNM stage and degree of tumor differentiation in HCC patients [63,64]. Fu et al. reported telomere length in serum cfDNA was significantly higher in the hepatitis B virus (HBV) related to HCC cases compared to controls [65]. DNA integrity has been shown associated with shorter overall survival, large tumor size, high TNM stage, presence of vascular and lymphatic invasion, and metastasis in HCC patients [66].
The presence of genetic mutations and epigenetic alterations to distinguish ctDNA from wild-type cfDNA provides a more specific biomarker for HCC diagnosis. However, the main issue with gene mutation detection in ctDNA from HCC is that the changes are highly varied, with few hot spots of frequent mutation. The tumor suppressor TP53 249Arg→Ser mutation is one exception, and it is frequently linked with aflatoxin exposure and HBV infection in HCC [67–69]. Higher levels of mutations in the human telomerase gene (hTERT) have also been reported in HCC patients with HCV and multinodular HCC [70].
DNA hypermethylation or hypomethylation changes frequently exist in cancerous cells which have a huge impact on tumor growth, leaving a specific gene signature. cfDNA methylation patterns have significant diagnostic and prognostic utility in HCC patients. Wong et al. were the first to report genes’ promoter methylation in HCC; in this case, they demonstrated that changes in methylation of promoters of both p15 and p16 genes are present in the blood samples of HCC patients [71,72]. Other studies of ctDNA showed that methylation of the promoter region in the Ras association domain family 1 domain A (RASSF1A) gene exists in 58% of HBV carriers and 93% of HCC patients [73]. RASSF1A, as well as p15 and p16, adenomatous polyposis coli (APC), fragile histidine triad (FHIT), and E-cadherin genes’ promoters methylation changes are usually high in ctDNA, a pivotal finding since changes may predate HCC diagnosis by up to 9 years. Zhang et al. demonstrated that the efficiency of RASSF1A, p15, and p16 genes methylation detection in ctDNA for HCC diagnosis was 89% [74,75]. Also, HCC patients have higher levels of INK4A Inhibitor gene promotor methylation in cfDNA [76]. Han et al. proved that hypermethylation of the TGR5 gene promoter in ctDNA was significantly more common in HCC cases than healthy controls [77]. Vaca-Paniagua et al. found that vimentin (VIM) gene promoter methylation was higher in ctDNA from HCC patients [78].
A variety of changes in methylation have been useful as diagnostic tools and to predict patient outcomes. HCC seropositive cases for the methylated Cyclin D2 gene exhibit a significantly shorter disease-free survival period than seronegative patients [79]. Hardy et al. showed that cfDNA methylation of the PPARγ gene promoter in a patient's plasma rises in correlation with hepatic fibrosis stage in both non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD) [80]. Moreover, a DNA methylation level test at this gene promoter sequence present in plasma outperforms the commonly used NAFLD fibrosis score, a test that helps to estimate the amount of scarring in the liver [81]. Hypomethylation of LINE1 DNA replicates in ctDNA serves as an independent predictor of reduced overall survival in HCC and is also linked with HBV etiology of liver disease, large tumor size, and advanced Cancer of the Liver Italian Program (CLIP) score [70]. Methylation of a single gene indicates limited value in the diagnosis of HCC, but by analyzing methylation of multiple genes (APC, RASSF1A, Glutathione S-transferase pi [GSTP1], and Secreted frizzled-related protein 1 precursor [SFRP1]). Huang et al. were able to improve the sensitivity to 92.7% and specificity to 81.9% [82]. In a related study, Xu et al., identified the ten most frequent methylation markers in cfDNA of HCC patients to configure a diagnostic model. They applied the model in a cohort of HCC cases and controls, finding that the model reached a sensitivity of 83.3% and a specificity of 90.5% [83]. While cfDNA has been most commonly isolated from the blood it is not restricted to that fluid compartment. It can also be isolated from urine and referred to as trans renal cfDNA (or Tr-DNA). A recent study showed that monitoring methylation changes in RASSF1A and GSTP1 genes from cfDNA in the urine samples following HCC resection allowed early detection of recurrence up to 9 months before the disease was detected by magnetic resonance imaging (MRI) [84].
4Cell-free messenger RNAStudies of cell-free messenger RNA (cfmRNA) have been hampered because it is readily degraded by ribonuclease(s) in the bloodstream and its quantity in plasma/serum is exceedingly small to be widely used as a diagnostic tool. However, the recent discovery of the presence of mRNAs incorporated into exosomes, microvesicles, and multi vesicles, has renovated the study of these particles’ mRNA species as disease biomarkers [85].
4.1Noncoding RNAAbout 20% of genomic DNA does not have a biochemical function; instead, it is transcribed into several noncoding RNAs (ncRNAs) that are very important for many cellular actions related to the synthesis of proteins from the genome coding regions [86,87]. There are two general types of ncRNAs based on size, long (IncRNAs) and short (SncRNAs) non-coding molecules, both of which are involved in gene expression regulation.
About 10 different IncRNAs are perceptible in HCC patients’ blood work. Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) and Sprouty Receptor Tyrosine Kinase Signaling Antagonist 4-Intronic Transcript 1 (SPRY4-1T1) IncRNAs levels increase with the grade of HCC and resection of the tumor leads to their decrease in the circulation [88,89]. Differences in plasma levels of the following lncRNAs (XLOC014172,LINC00152, and RP11-160H22.5) can distinguish HCC from chronic hepatitis, cirrhosis or normal liver [90].
For circulating SncRNAs, miRNAs have generated special attention, these are generated in the bloodstream both by active secretion and cell lysis [91]. A given miRNA may regulate gene expression to control several cellular processes, including development, differentiation, metabolism, and cell death; in some cases, specific miRNAs appear to function like oncogenes or tumor suppressor genes [92,93] and in HCC more than 70 miRNAs found in the circulation have been suggested to be useful as biomarkers. Li et al. illustrated that miRNAs serum profiling work as biomarkers to discriminate between HBV infection and HBV-positive HCC [94]. Nakamura et al. showed that the circulating miR-122 level in combination with Wisteria floribunda agglutinin-positive Mac-2 binding protein achieves 47% sensitivity and 87% specificity in detecting advanced fibrosis related to HBV infection [95]. High serum levels of miR-221 have also been linked to tumor size, degree of cirrhosis, and tumor stage in HCC patients [96] (Table 1).
Circulating cell-free microRNA in hepatocellular carcinoma cases.
| miRNA | Expression | HCC | Reference |
|---|---|---|---|
| miR-1 | Up-regulated | 195 | Koberle et al. [97] |
| miR-10b | Up-regulated | 27 | Jiang et al. [98] |
| miR-15b | Up-regulated | 153 | Yin et al. [8] |
| miR-15b-5p | Downregulated | 37 | Chen et al. [99] |
| miR-16 | Downregulated | 105 | Yin et al. [8] |
| miR-17-5p | Up-regulated | 136 | Zheng et al. [95] |
| miR-18a | Up-regulated | 101 | Shen et al. [100] |
| miR-18b | Up-regulated | 51 | Rashad et al. [101] |
| miR-19a | Downregulated | 112 | Motawi et al. [102] |
| miR-20a | Up-regulated | 150 | Fahim et al. [103] |
| miR-21 | Up-regulated | 457 | Shen et al. [100] |
| Downregulated | 70 | Qi et al. [104] | |
| miR-22 | Downregulated | 192 | Zekri et al. [105] |
| miR-23b-3p | Downregulated | 40 | Sun et al. [106] |
| miR-24-3p | Up-regulated | 84 | Shen et al. [100] |
| miR-26a | Downregulated | 457 | Shen et al. [100] |
| miR-26a-5p | Downregulated | 261 | Tan et al. [107] |
| miR-27a | Downregulated | 457 | Shen et al. [100] |
| Up-regulated | 51 | Rashad et al. [101] | |
| miR-29b | Downregulated | 192 | Zekri et al. [105] |
| miR-30c | Downregulated | 242 | Liu et al. [108] |
| miR-30c-5p | Downregulated | 8 | Shen et al. [100] |
| miR-34a | Up-regulated | 112 | Motawi et al. [102] |
| miR-92a-3p | Up regulated | 20 | Yin et al. [8] |
| miR-96 | Up-regulated | 104 | Chen et al. [109] |
| miR-99a | Downregulated | 30 | Ning et al. [110] |
| miR-101 | Up-regulated | 25 | Fu et al. [111] |
| Downregulated | 67 | Shen et al. [100] | |
| miR-101-3p | Up-regulated | 38 | Moshiri et al. [112] |
| miR-106b | Up-regulated | 27 | Jiang et al. [98] |
| miR-106b-3p | Up-regulated | 38 | Moshiri et al. [112] |
| miR-122 | Up-regulated | 192 | Zekri et al. [105] |
| Downregulated | 457 | Shen et al. [100] | |
| miR-122a | Downregulated | 85 | Luo et al. [113] |
| miR-122-5p | Up-regulated | 120 | Shen et al. [100] |
| Downregulated | 261 | Tan et al. [107] | |
| miR-125a-5p | Up-regulated | 20 | Oura et al. [114] |
| miR-125b | Downregulated | 64 | Chen et al. [122] |
| miR-125b-5p | Up-regulated | 20 | Yin et al. [8] |
| miR-126 | Up-regulated | 49 | Ghosh et al. [116] |
| Downregulated | 23 | Khairy et al. [117] | |
| miR-128-2 | Up-regulated | 222 | Zhuang et al. [118] |
| miR-129 | Downregulated | 23 | Khairy et al. [117] |
| miR-130a | Up-regulated | 112 | Motawi et al. [102] |
| miR-130b | Up-regulated | 153 | Yin et al. [8] |
| miR-132 | Downregulated | 80 | Wang et al. [119] |
| miR-139 | Downregulated | 31 | Shen et al. [100] |
| miR-141-3p | Up-regulated | 261 | Tan et al. [107] |
| miR-143 | Up-regulated | 95 | Yin et al. [8] |
| miR-143-3p | Up regulated | 49 | Ghosh et al. [116] |
| miR-146a | Up-regulated | 112 | Motawi et al. [102] |
| miR-148a | Downregulated | 155 | Han et al. [120] |
| miR-150 | Downregulated | 120 | Shen et al. [100] |
| miR-155 | Downregulated | 23 | Khairy et al. [117] |
| Up-regulated | 30 | Ning et al. [110] | |
| miR-181a | Downregulated | 27 | Jiang et al. [98] |
| miR-181b | Up-regulated | 192 | Zekri et al. [105] |
| miR-182 | Up-regulated | 103 | Shen et al. [100] |
| miR-192 | Up-regulated | 457 | Shen et al. [100] |
| miR-192-5p | Downregulated | 261 | Tan et al. [107] |
| miR-195 | Downregulated | 112 | Motawi et al. [102] |
| miR-199a | Downregulated | 105 | Yin et al. [8] |
| miR-199a-3p | Downregulated | 192 | Zekri et al. [105] |
| miR-199a-5p | Downregulated | 261 | Tan et al. [107] |
| miR-200a | Up-regulated | 136 | Liu et al. [121] |
| miR-203 | Downregulated | 23 | Khairy et al. [117] |
| miR-203a | Downregulated | 242 | Liu et al. [108] |
| miR-206 | Up-regulated | 261 | Tan et al. [107] |
| miR-212 | Downregulated | 80 | Wang et al. [119] |
| miR-215 | Up-regulated | 95 | Yin et al. [8] |
| miR-218 | Downregulated | 156 | Yang et al. [122] |
| miR-221 | Up-regulated | 192 | Zekri et al. [105] |
| miR-222 | Up-regulated | 70 | Qi et al. [104] |
| miR-223 | Up-regulated | 101 | Shen et al. [100] |
| Downregulated | 457 | Shen et al. [100] | |
| miR-223-3p | Downregulated | 20 | Yin et al. [8] |
| miR-224 | Up-regulated | 122 | Lin et al. [123] |
| miR-224-5p | Up regulated | 136 | Liu et al. [121] |
| miR-296 | Up-regulated | 112 | Motawi et al. [102] |
| miR-302c-3p | Downregulated | 8 | Shen et al. [100] |
| miR-331-3p | Up-regulated | 103 | Shen et al. [100] |
| miR-335 | Downregulated | 125 | Cui et al. [124] |
| miR-338-5p | Up-regulated | 37 | Chen et al. [99] |
| miR-375 | Up-regulated | 120 | Shen et al. [100] |
| Downregulated | 78 | Shen et al. [100] | |
| miR-433-3p | Up-regulated | 261 | Tan et al. [107] |
| miR-483-5p | Up-regulated | 112 | Zhang et al. [125] |
| miR-500a | Up-regulated | 112 | Zhang et al. [125] |
| miR-574-3p | Up-regulated | 70 | Shen et al. [126] |
| miR-764 | Up-regulated | 37 | Chen et al. [99] |
| miR-801 | Up-regulated | 457 | Shen et al. [100] |
| miR-885-5p | Up regulated | 192 | Zekri et al. [105] |
| miR-1228-5p | Up regulated | 261 | Tan et al. [107] |
| miR-1246 | Up-regulated | 38 | Moshiri et al. [112] |
| miR-1291 | Up-regulated | 50 | Hagag et al. [127] |
| let-7b | Up-regulated | 120 | Shen et al. [100] |
HCC: Hepatocellular carcinoma.
Measurements of miRNAs have also been studied in the context of therapeutic monitoring. Zheng et al. concluded that the serum levels of miR-17-5p could be a prognostic marker for HCC patients past their surgical resection, and Cho et al. showed that patients with HBV-related HCC who underwent radiofrequency ablation (RFA) had low overall survival rates if associated with high plasma miR-122 expression [100,125]. Yamamoto et al. indicated that miR-500 is an oncofetal miRNA found abundantly in the serum of HCC patients, levels return to normal after successful surgery [126]. For unresectable HCC, Liu et al. displayed that miR-200a was an independent prognostic factor linked to survival rate after trans-arterial chemoembolization (TACE) [121]. PTEN, Stathmin1, RUNX3, Rho-kinase 2, Mcl-1, SOX9, FNDC3B, p21/E2F5, VEGF, TP53INP1, ADAM17, ISRE, CDKN1B/p27, CDKN1C/p57, TIMP3, HDAC4, mTOR, and LIN28B have cancer-related functions, certain miRNAs in patients with HCC are targeted by these [98,8,100,102,115,121,127].
In attempts to improve the accuracy of diagnosis, some researchers have measured multiple miRNAs in serum. Zekri et al. could differentiate HCC from healthy, chronic hepatitis B, and cirrhosis by measuring the levels of 7 miRNAs (miR-122, -192, -21, -223, -26a, -27a, and -801) in plasma [100]. More recently, a panel of miR-29b, miR-122, and miR-885 in combination with AFP showed high diagnostic accuracy in detecting HCC within the normal population, while a combination of AFP with miR-22, miR-122, miR-221, and miR-885 was superior at diagnosing of HCC with a background of cirrhosis [128].
In acute liver injury and hepatitis, circulating miRNAs are regulated by intrahepatic oxidative stress, and seem to determine the extent of the damage. miR-122-5p is profusely expressed in hepatocytes and increases in the serum after excessive alcohol intake [129]. Mosedale et al. reported early increases of miR-122-5p are associated with mitochondrial-induced apoptosis and oxidative stress during drug-induced liver injury [130] (Table 2).
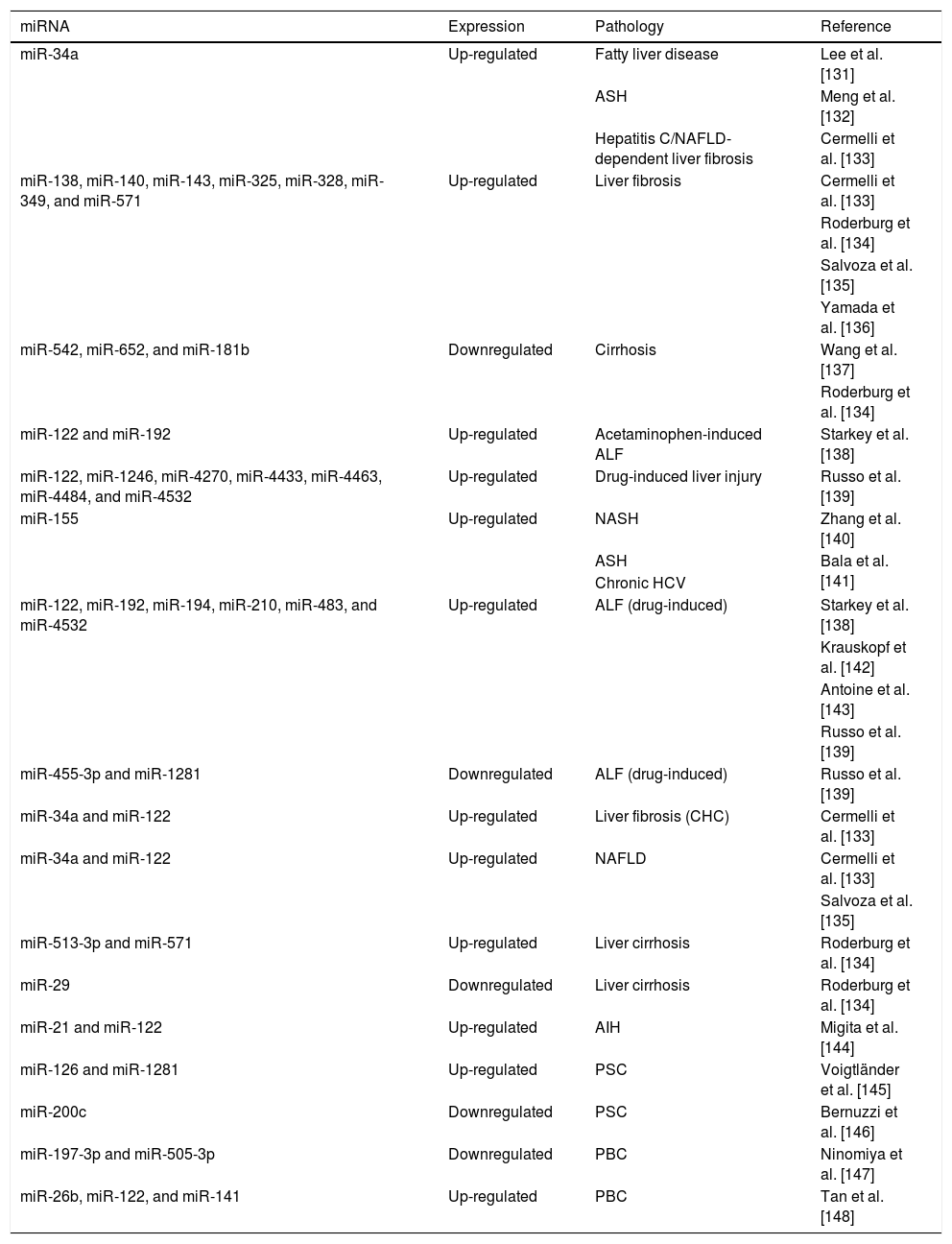
Circulating cell-free microRNA in liver diseases.
| miRNA | Expression | Pathology | Reference |
|---|---|---|---|
| miR-34a | Up-regulated | Fatty liver disease | Lee et al. [131] |
| ASH | Meng et al. [132] | ||
| Hepatitis C/NAFLD-dependent liver fibrosis | Cermelli et al. [133] | ||
| miR-138, miR-140, miR-143, miR-325, miR-328, miR-349, and miR-571 | Up-regulated | Liver fibrosis | Cermelli et al. [133] |
| Roderburg et al. [134] | |||
| Salvoza et al. [135] | |||
| Yamada et al. [136] | |||
| miR-542, miR-652, and miR-181b | Downregulated | Cirrhosis | Wang et al. [137] |
| Roderburg et al. [134] | |||
| miR-122 and miR-192 | Up-regulated | Acetaminophen-induced ALF | Starkey et al. [138] |
| miR-122, miR-1246, miR-4270, miR-4433, miR-4463, miR-4484, and miR-4532 | Up-regulated | Drug-induced liver injury | Russo et al. [139] |
| miR-155 | Up-regulated | NASH | Zhang et al. [140] |
| ASH | Bala et al. [141] | ||
| Chronic HCV | |||
| miR-122, miR-192, miR-194, miR-210, miR-483, and miR-4532 | Up-regulated | ALF (drug-induced) | Starkey et al. [138] |
| Krauskopf et al. [142] | |||
| Antoine et al. [143] | |||
| Russo et al. [139] | |||
| miR-455-3p and miR-1281 | Downregulated | ALF (drug-induced) | Russo et al. [139] |
| miR-34a and miR-122 | Up-regulated | Liver fibrosis (CHC) | Cermelli et al. [133] |
| miR-34a and miR-122 | Up-regulated | NAFLD | Cermelli et al. [133] |
| Salvoza et al. [135] | |||
| miR-513-3p and miR-571 | Up-regulated | Liver cirrhosis | Roderburg et al. [134] |
| miR-29 | Downregulated | Liver cirrhosis | Roderburg et al. [134] |
| miR-21 and miR-122 | Up-regulated | AIH | Migita et al. [144] |
| miR-126 and miR-1281 | Up-regulated | PSC | Voigtländer et al. [145] |
| miR-200c | Downregulated | PSC | Bernuzzi et al. [146] |
| miR-197-3p and miR-505-3p | Downregulated | PBC | Ninomiya et al. [147] |
| miR-26b, miR-122, and miR-141 | Up-regulated | PBC | Tan et al. [148] |
AIH: Autoimmune hepatitis, ALF: Acute liver failure, ASH: Alcoholic steatohepatitis, HCV: Hepatitis C virus, NAFLD: Non-alcoholic fatty liver disease, NASH: Non-alcoholic steatohepatitis, PBC: Primary biliary cholangitis, PSC: Primary sclerosing cholangitis.
Cells liberate extracellular vesicles (EVs) into the extracellular environment either as part of normal physiological functions or during pathological processes. Their function is communication between cells. EVs contents (mRNAs, miRNAs, proteins, and lipids) indicate the cell of origin as well as the process or stress that activates their formation and release [149,150]. EVs are classified based on their size and biogenesis: exosomes or ectosomes (or microparticles [MPS]) [151]. A growing number of studies have provided data for a key pathophysiological part of EVs in various aspects of liver injury [152,153]. These findings, in combination with the fact that EVs are released into the systemic circulation and are singularly stable in this environment, support the concept that assessment and quantitation of EVs in blood might provide novel information from the liquid biopsy with liver disease (see Fig. 1).
MPS levels in the blood are altered in certain disease states. Kornek et al. witnessed an increment in MPs enriched in the surface markers from monocytes and natural killer T cells and a decline in MPs collected from neutrophils and leuco-endothelial cells in NAFLD patients [154,155]. Rautou et al. reported that MPs enriched in leuco-endothelial cell markers were higher in patients with cirrhosis in comparison with healthy controls, and were increasingly elevated with the severity of cirrhosis assessed using Child–Pugh scores [156]. A limitation of measuring MPS levels is that myeloid cell-derived EVs are discharged into the bloodstream in a wide variety of inflammatory conditions and are thus not specific for hepatic disease. To improve the specificity, many groups have sought to identify liver-specific protein markers such as Asialoglycoprotein Receptor (ASGPR1), Cytokeratin-18, and vanin-1 in blood EVs and quantitate their levels [157–161]. Mann et al., and Arbelaiz et al., examined the proteomes of circulating EVs in NAFLD, non-alcoholic steatohepatitis (NASH), primary sclerosing cholangitis (PSC), cholangiocarcinoma (CCA), and HCC, and found multiple differences in the proteins expressed in EVs in the different patient groups [151,162]. Unfortunately, the diagnostic value of the various protein levels was not analyzed, and future studies are needed to determine whether proteomic signatures in serum EVs can be developed as a useful diagnostic tool for patients with CCA, PSC, and HCC.
Lately, the role of the microbiome has acquired attention as a contributor to the development of liver diseases [163]. Molecular technology has recognized the existence of microbiota in the bloodstream by sequencing 16S rDNA bacterial genes [164]. The DNA is extracted out of EVs, and then the bacterial genomic DNA is amplified. Puri et al. reported a decrease in Bacteroidetes, and an increase in Fusobacteria, in patients with heavy alcohol consumption [165]. Also, Cho et al. reported a decrease in Pseudomonas, Streptococcus, and Bifidobacterium, and an increase in Staphylococcus, Acinetobacter, Klebsiella, and Trabulsiella, associated with HCC patients [166].
Besides proteins, other EV contents of interest are miRNAs. miRNAs are dropped into the extracellular space and bloodstream where they are protected from degradation either by association with proteins such as those from the Ago2 family or their presence within EVs. In the context of hepatic disease, particular attention has been concentrated on miRNA-122 and miRNA-192 due to their tissue specificity in the liver where they represent over 70% of the total miRNA population [167]. Pirola et al. indicated that in healthy people miR-122 is available in the circulation only in an Ago2 complex, but in patients with NAFLD, most miR-122 in serum exists in Ago2-free forms [168]. Bala et al. showed that miRNA-122 is elevated in different types of liver disease but is found in different forms; predominantly in EVs in ALD and protein-enriched fractions in the bloodstream in acetaminophen-induced liver injury [141].
Using an untargeted approach of profiling the miRNA content from EVs released by hepatocytes cut off from a gastric infusion model of ALD, Eguchi et al., demonstrated that, in experimental alcoholic steatohepatitis (ASH) mice, there is an increment generation and release of EVs that enclose a miRNA signature detectable in blood EVs as a barcode to recognize ASH. ALD is a wide-spectrum pathology that includes steatosis, steatohepatitis, and in severe cases, fibrosis and/or cirrhosis. Their study labeled 13 significantly up- or down-regulated miRNAs in ASH EVs compared to control EVs. Eight miRNAs from the nine significantly up-regulated in ASH EVs (miR-541, miR-3473, miR-143, miR-29a, let-7f, miR-340, miR-34a, and miR-3473b) were implicated in inflammatory and cancer pathways and potentially regulate 121 objective genes, including Bcl2, JUN, IL-6, PTEN, SMAD, and Wnt families. In a further small human study, consisting of a group of ALD patients and nonalcoholics controls, the results demonstrated a significant increment in blood EVs in patients with ALD. Besides, the identified profile of three miRNAs (let-7f, miR-29, and miR-340) was significantly higher in these patients examined in contrast to nonalcoholics [169].
In summary, growing evidence is pointing to the potential role of EVs as a novel liquid biopsy for liver disease approach and opens the possibility of a new era of precision medicine where disease-specific and liver injury-specific signatures can be accurately identified in a non-invasive manner. However, future studies addressing the importance of changes the levels of circulating miRNAs, their compartmentalization in EVs, and/or association with serum proteins, and the differences between levels, packaging in EVs and protein complexes in different forms of liver diseases are needed to assess potential diagnostic and prognostic utility.
6ConclusionsLiquid biopsy has several advantages above standards clinical tools, providing specific, dynamic and fast access information to different illnesses. In the case of CLD, including HCC, it is a promising diagnostic, therapeutic and prognostic tool, leading to being a fundamental part of precision medicine.
Different biomarkers can be extracted from the liquid biopsy. In the case of CECs, there are limitations regarding the isolation techniques, physical or biological. The most used are the biological methods, which depend on the antigen-antibody binding. Until now, the most standardized process involves EpCAM, with evidence of usefulness in the diagnosis, response to treatment and recurrence of HCC patients. However, alternative biomarkers are emerging with greater sensitivity and specificity, whose detection is carried out using chips, improving the purification rate.
Concerning cfDNA in HCC patients, there is an increase in cases of metastasis progression and worse prognosis, although this type of change is not disease-specific. However, more specific genetic mutations and epigenetic alterations for HCC have been found. The most important qualitative change is in the methylation pattern, showing a significant association of methylation features of p15, p16 and RASSF1A genes with HCC, and of the PPARγ and LINE1 loci with benign liver diseases.
miRNAs are generated by the cell active secretion or by cellular lysis. Up to know, around 70 miRNAs are related to HCC, which function as a diagnostic and treatment monitoring tool. More recently, panels have been developed, that include some miRNAs increasing the sensitivity and specificity for different CLD.
EVs contain different cellular products, which make it very useful as a tool by having different types of approaches in the patient. In the case of the CLD, some proteins and miRNAs present in EVs have been analyzed, finding interesting results that require further research to prove their usefulness.
Liver damage, through its main causes, i.e. alcohol, virus, and obesity can be detected at an early stage through the liquid biopsy, which has been proving to be a reliable and convenient tool in the new era of genomic medicine in hepatology.AbbreviationsCLD chronic liver diseases hepatocellular carcinoma circulating free DNA circulating epithelial cells circulating tumor cells isolation by size of epithelial tumor cells circulating tumor microemboli epithelial cell adhesion molecule human epidermal growth factor receptor prostate-specific antigen Food and Drug Administration epithelial-mesenchymal transition hydroxyapatite/chitosan aptamer for carbohydrate sialyl Lewis X real-time polymerase chain reaction alfa fetoprotein melanoma-associated antigens glypican protein intracellular adhesion molecule neutrophil extracellular traps glycoprotein circulating free tumor DNA hepatitis C virus hepatitis B virus human telomerase gene Ras association domain family 1 domain A adenomatous polyposis coli fragile histidine triad vimentin non-alcoholic fatty disease liver alcoholic liver disease cancer of the Liver Italian Program glutathione S-transferase pi secreted frizzled-related protein 1 precursor trans renal circulating free DNA magnetic resonance imaging cell-free messenger RNA noncoding RNAs long noncoding RNAs short noncoding RNAs Metastasis Associated Lung Adenocarcinoma Transcript 1 Sprouty Receptor Tyrosine Kinase Signaling Antagonist 4-Intronic Transcript 1 microRNAs radiofrequency ablation trans-arterial chemoembolization extracellular vesicles microparticles Asialoglycoprotein Receptor 1 non-alcoholic steatohepatitis primary sclerosing cholangitis cholangiocarcinoma alcoholic steatohepatitis
The authors have no conflicts of interest to declare.
Thanks to Rolando H. Gonzalez for critical reading of the manuscript.






![(A) Liquid biopsy of hepatocellular carcinoma (HCC): Spontaneous circulation of CTCs and CTM in peripheral blood reflects tumor progression and tumor spread in patients with HCC [20]. (B) Cell-free nucleic acids (cfNA): cfNA (DNA and RNA) are known to come from apoptotic and necrotic cells or to be released from living eukaryotic cells, thereby providing a valuable source of material which can instruct about natural or biological and pathological processes within its cellular source [170]. (C) Extracellular vesicles (EVs): EVs are small membrane vesicles released by cells in the extracellular environment as part of normal physiology or during pathological processes, which function is the communication between cells, their cargo (mRNAs, miRNAs, proteins and lipids) may reflect the cell of origin as well as the specific stress that induces their formation and release [149,150]. Red blood cell (RBC), white blood cell (WBC), circulating tumor cells (CTCs), circulating tumor microemboli (CTM), and cell-free DNA (cfDNA). (A) Liquid biopsy of hepatocellular carcinoma (HCC): Spontaneous circulation of CTCs and CTM in peripheral blood reflects tumor progression and tumor spread in patients with HCC [20]. (B) Cell-free nucleic acids (cfNA): cfNA (DNA and RNA) are known to come from apoptotic and necrotic cells or to be released from living eukaryotic cells, thereby providing a valuable source of material which can instruct about natural or biological and pathological processes within its cellular source [170]. (C) Extracellular vesicles (EVs): EVs are small membrane vesicles released by cells in the extracellular environment as part of normal physiology or during pathological processes, which function is the communication between cells, their cargo (mRNAs, miRNAs, proteins and lipids) may reflect the cell of origin as well as the specific stress that induces their formation and release [149,150]. Red blood cell (RBC), white blood cell (WBC), circulating tumor cells (CTCs), circulating tumor microemboli (CTM), and cell-free DNA (cfDNA).](https://static.elsevier.es/multimedia/16652681/000000200000000C/v2_202101071056/S1665268120300387/v2_202101071056/en/main.assets/thumbnail/gr1.jpeg?xkr=ue/ImdikoIMrsJoerZ+w96p5LBcBpyJTqfwgorxm+Ow=)