Deregulation of m6A methylation, the most prevailing RNA modification, participates in cancer pathogenesis. METTL16, an atypical methyltransferase, functions as a pro-tumorigenic factor in hepatocellular carcinoma (HCC). Here, we explored the action of METTL16 on HCC glycolysis and the associated mechanism.
Materials and MethodsExpression analysis was done by quantitative PCR, immunoblotting, or immunohistochemistry. Cell sphere formation, invasiveness, apoptosis, proliferation and viability were detected by sphere formation, transwell, flow cytometry, EdU and CCK-8 assays, respectively. Xenograft studies were performed to analyze the role in vivo. Methylated RNA immunoprecipitation (MeRIP) and RIP assays were used to verify the METTL16/PFKM relationship. PFKM mRNA stability was tested by actinomycin D treatment. Chromatin immunoprecipitation (ChIP) and luciferase assays were performed to analyze the POU3F2/METTL16 relationship.
ResultsIn HCC, METTL16 expression was elevated, and increased levels of METTL16 transcript predicted poor HCC prognosis. METTL16 deficiency resulted in suppressed HCC cell growth, invasiveness and sphere formation. Moreover, METTL16 depletion diminished HCC cell glycolysis. Mechanistically, PFKM expression was positively associated with METTL16 expression. METTL16 mediated m6A methylation to stabilize PFKM mRNA via an IGF2BP3-dependent manner. Restored PFKM expression exerted a counteracting effect on METTL16 deficiency-mediated in vitro cell phenotype alterations and in vivo xenograft growth suppression. Furthermore, POU3F2 promoted the transcription of METTL16 in HCC cells.
ConclusionsOur findings define the crucial role of the POU3F2/METTL16/PFKM axis in HCC pathogenesis, offering the potential opportunity to combat HCC.
Liver cancer leads to more a million cancer-associated deaths (7.8 % of the total cancer deaths) throughout the world in 2022 and has become the third predominant determinant of cancer mortality, with hepatocellular carcinoma (HCC) accounting for 75–85 % of all cases [1]. The etiology of HCC is closely linked to multiple factors, such as hepatitis virus infection, chronic liver disease, and gene susceptibility [2]. Despite the advantage of advances of systemic therapies in the early stage, HCC remains terrible survival because it is often confirmed at an advanced stage [2,3]. Currently, studies exploring the molecular pathogenesis of HCC are needed to devise novel markers for HCC diagnosis and management at all stages. Enhanced glycolysis can facilitate cancer cell survival and contribute to pro-cancerous tumor environment. Targeting glycolysis has been proposed to be useful for the improvement of HCC therapy [4].
Deregulation of m6A methylation, the most prevailing RNA modification in eukaryotes, has been reported in human cancer [5]. m6A modification is controlled by methyltransferases (m6A writers), demethylases (m6A erasers) and m6A binding proteins (m6A readers). METTL16, a member of methyltransferase complex, is capable of functioning as an atypical methyltransferase to catalyze RNA m6A methylation, thereby actively participating in cancer pathogenesis [6]. For instance, METTL16-mediated m6A modification contributes to leukemogenesis by affecting branched-chain amino acid (BCAA) metabolism by upregulating the levels of BCAA transaminase 1 and 2 [7]. METTL16 enhances cyclin D1 mRNA m6A modification to stabilize the transcript, thus leading to enhanced proliferation of gastric cancer cells [8]. The pro-tumorigenic role of METTL16 in HCC has begun to emerge; for example, METTL16 accelerates HCC development by affecting the stabilization of RAB11B-AS1 transcript via an m6A mechanism [9]; METTL16 mediates m6A methylation of Lnc-CSMD1–7 to induce HCC metastasis [10]. Nonetheless, our understanding of the action of METTL16 on HCC glycolysis and its associated mechanism has remained incomplete.
In the current work, our data show new evidence for the pro-tumorigenic role of METTL16 in HCC. We further demonstrate the promoting activity of METTL16 in HCC glycolysis. Through bioinformatic and experimental analyses, we elucidated an uncharacterized associated mechanism, providing a novel foundation for targeting METTL16 as an encouraging target for HCC management.
2Materials and Methods2.1BioinformaticsThe previously developed ENCORI algorithm (https://rnasysu.com/encori/) was used to observe the expression pattern of METTL16 transcript in HCC, to reveal the expression correlation of PFKM with METTL16 or IGF2BP3 in HCC, and to predict m6A readers associated with PFKM mRNA. The open-access TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga) was used for analysis of PFKM or IGF2BP3 transcript expression in HCC. To evaluate the association of HCC overall survival with METTL16, IGF2BP3 or PFKM expression, the online tool Kaplan-Meier Plotter was used at https://www.kmplot.com/analysis/. We applied the human protein atlas online resource (https://www.proteinatlas.org/) to analyze METTL16 protein expression in HCC. The GSE224008 dataset on NCBI (https://www.ncbi.nlm.nih.gov/) was applied to reveal differentially expressed genes (DEGs) after METTL16 silencing in Huh-7 liver cancer cells, and these DEGs were subjected to KEGG pathway enrichment analysis at https://www.bioinformatics.com.cn/?keywords=pathway. We used the online website SRAMP to predict the putative m6A methylation sites of PFKM mRNA at http://www.cuilab.cn/sramp. To search for the transcription factors (TFs) in promoting METTL16 transcription, we utilized three TF-predicting algorithms Genecards (https://www.genecards.org/), PROMO (https://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) and JASPAR (https://jaspar.elixir.no/). The JASPAR algorithm was also used to predict the putative binding sites for POU3F2 in the METTL16 promoter.
2.2Human tissue specimensIn this study, we obtained fresh-frozen human primary HCC tumors (n = 40) and matched non-cancerous tissues (n = 40) from HCC patients from the First Affiliated Hospital, Hengyang Medical School, University of South China, after acquisition of patients’ informed consent. Clinical follow-up ranged from 6 to 60 months. Human research protocols were authorized by the First Affiliated Hospital, Hengyang Medical School, University of South China Ethics and Scientific Committee. In accordance with the standard guidelines, we extracted total RNA and protein from these specimens.
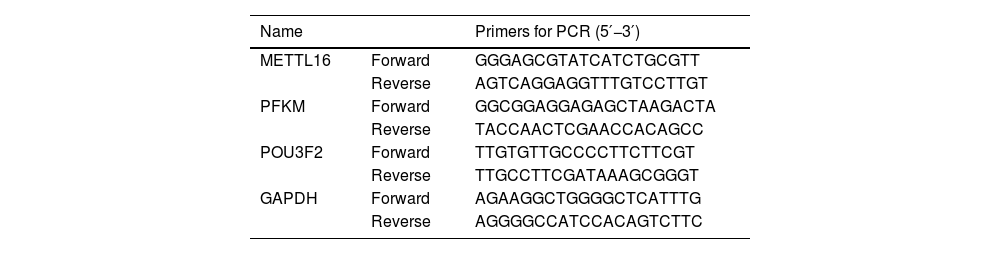
2.3mRNA expression by quantitative PCRUsing the BeyoMag™ RNA Kit and accompanying suggestions (Beyotime, Shanghai, China), we prepared total RNA from human tissues. 500 ng of RNA was subjected to reverse transcription using the QuantiTect RT Kit as per the manufactory suggestions (Qiagen, Tokyo, Japan). To gauge the levels of POU3F2, METTL16, and PFKM transcripts, quantitative PCR was done following cDNA generation under the application of SYBR Green mix (Takara, Dalian, China) and gene-specific primers (Table 1). For relative quantification analysis, GAPDH served as a reference gene, and the 2−ΔΔCt method was used.
2.4Cell linesThis study included MHCC97 (#C6585, Beyotime), Huh-7 (#IM-H040, Immocell, Xiamen, China), and Hep3B HCC cells (#IM-H367, Immocell) and THLE-2 cells (#CL-0833, Procell, Wuhan, China). For cell cultivation (MHCC97, Huh-7, Hep3B), we utilized the complete medium consisted of 89 % DMEM, 10 % FBS and 1 % streptomycin/penicillin (all from Beyotime). THLE-2 cells were propagated in Human liver immortalized cell growth media purchased from Procell. A humidified 5 % CO2 incubator (Thermo Fisher Scientific, Geel, Belgium) was used for cell culture at 37 °C.
2.5SiRNAs, plasmids, lentivirus particles, transfection, and transductionThis study used the following siRNAs (MedChemExpress, Shanghai, China): si-METTL16#1, si-METTL16#2, si-METTL16#3, si-IGF2BP3, si-YY1, si-POU3F2, and si-NC control. The 1:1 ratio mix of si-METTL16#1 and si-METTL16#3 was named as si-METTL16. pCMV-IGF2BP3(human)−3×FLAG-Neo, pCMV-3×FLAG-PFKM(human)-Neo, and matched control vector were procured from Miaoling Biology (Wuhan, China). Lentivirus particles expressing sh-METTL16 or sh-NC control were from Genomeditech (Shanghai, China).
For transfection, we seeded MHCC97 and Huh-7 HCC cells 18–20 h before transfection to yield 60–75 % confluence at the time of transfection. Using Lipofectamine 3000 as suggested by the supplier (Thermo Fisher Scientific), liposomal cocktails with siRNA or/and plasmid were prepared in Opti-MEM (Thermo Fisher Scientific). Transfection complexes were added into cells, followed by incubation for 8 h before media change. For the further use, we obtained transfected cells that were cultured for 24–72 h.
For viral transduction, we plated 1 × 106 MHCC97 cells into a 10-cm dish 10–12 h prior to infection of sh-METTL16 or sh-NC lentivirus for 18–24 h Following media change, cells were subjected to drug selection (2 µg/mL puromycin) for at least 14 days.
2.6Animal studies, immunohistochemistry, and H&E stainingMouse xenograft studies were conducted following protocols approved by the First Affiliated Hospital, Hengyang Medical School, University of South China Animal Care and Use Ethics Committee Hospital. For these studies, we used female BALB/c nude mice (42–56-day old, n = 24, Vital River Laboratory, Beijing, China) and divided them in three groups: sh-NC (n = 8), sh-METTL16 (n = 8), and sh-METTL16+PFKM (n = 8). For subcutaneous xenograft generation, 100 µL of suspension containing 1.5 × 106 lentivirus-infected MHCC97 cells was mixed with 100 µL of Matrigel (BD Biosciences, Stockholm, Sweden) prior to cell implantation in nude mice by subcutaneous injection (n = 5 for each group). For metastasis analysis, MHCC97 cells were intravenously into nude mice through tail vein injection (n = 3 for each group). Intratumoral or nasal administration of the PFKM expression lentivirus (107 TU/mouse) was performed on day 8. Xenograft tumors were gauged every three days by caliper (volume=longest diameter × (shortest diameter)2 × 0.5) until resected. Mice were used for lung collection (metastatic node quantification) at week 6.
A portion of xenograft tumors were inflated into 4 % paraformaldehyde, embedded in paraffin, sectioned (4–5 µm), and subjected to immunohistochemistry for probing of Ki67 and PFKM as described elsewhere [11], using rabbit anti-Ki67 pAb (#27309-1-AP, Proteintech, Wuhan, China, 1 to 5000) and rabbit anti-PFKM pAb (#ab237545, Abcam, Cambridge, UK, 1 to 150). Mouse lungs were fixed, embedded in paraffin, sectioned (4–5 µm), and subjected to H&E staining using a commercial staining kit (Beyotime).
2.7Protein analysis by immunoblottingWe obtained total protein from collected tissue specimens or cultured cell lines and performed western blot assays as reported [12]. For immunoblot analysis, we used mouse anti-POU3F2 mAb (#ab243045, Abcam, 1 to 1000), rabbit anti-METTL16 pAb (#19924-1-AP, Proteintech, 1 to 8000), rabbit anti-PFKM pAb (#ab237545, Abcam, 1 to 150), rabbit anti-IGF2BP3 pAb (#14642-1-AP, Proteintech, 1 to 6000), rabbit anti-YY1 mAb (#ab109237, Abcam, 1 to 5000), and mouse anti-β-actin mAb (#ab8226, Abcam, 1 to 1000).
2.8Cell viability and proliferation assaysIn a 96-well plate, MHCC97 and Huh-7 cells were subjected to transfection with si-NC, si-METTL16#1, si-METTL16#3, si-METTL16 (the 1:1 ratio mix of si-METTL16#1:si-METTL16#3), si-METTL16+vector or si-METTL16+PFKM expression plasmid. After 48 h, we utilized the CCK-8 Viability Assay Kit (Yeasen, Shanghai, China) to evaluate the number of viable cells and the EdU incorporation Assay Kit (Ribobio, Guangzhou, China) for cell proliferation analysis as recommended by the vendors. A Multiskan Ascent 354 reader (Thermo Fisher Scientific) was used for absorbance measurement at 450 nm. For fluorescence image acquisition, we utilized the FV3000 confocal microscope (Olympus, Tokyo, Japan).
2.9Flow cytometryMHCC97 and Huh-7 cells after 72 h transfection were subjected to staining by the Annexin V-FITC/PI Apoptosis Assay Kit as described by the supplier (BD Biosciences). For FACS analysis, we applied a Calibur flow cytometer with CellQuest software (BD Biosciences).
2.10Transwell invasion assayWe plated 7 × 104 MHCC97 and Huh-7 cells (in non-serum media) at 24 h post transcription atop each transwell pre-coated with Matrigel (BD Biosciences). 10 % FBS media was used as a chemoattractant in the bottom well. Following incubation for 24 h at 37 °C, the number of the cells remaining on the lower surface was quantified after crystal violet (1 %) staining.
2.11Sphere formation assayFor the assay, we used a 96-well Clear Round Bottom Ultra Low Attachment plate (Corning, Shanghai, China). We seeded 200 MHCC97 and Huh-7 cells after 24 h introduction into each well in non-serum media. Under standard protocols, cell culture was done for 7 days. We analyzed the efficiency of sphere formation by microscopic images.
2.12Detection of glucose consumption level, lactate production content, and ATP/ADP ratioMHCC97 and Huh-7 cells at 48 h post transfection were harvested for these detections. In accordance with the suppliers’ suggestions, the glucose consumption Assay Kit (Beyotime) was applied for glucose consumption content measurement, the lactate production Assay Kit (Abcam) was used to gauge lactate production level, and the ATP/ADP ratio Chemiluminescence Assay kit (Elabscience, Wuhan, China) was employed to analyze the ATP/ADP ratio.
2.13Methylated RNA immunoprecipitation (MeRIP) and RIP assaysSi-METTL16-transfected, si-NC-introduced, and untransfected MHCC97 and Huh-7 cells were subjected to RIP experiments using BeyoRIP™ RIP Assay Kit as recommended by the producer (Beyotime). The complex of Protein A/G Agarose and rabbit anti-m6A mAb (#MA5-33030, Thermo Fisher Scientific), rabbit anti-METTL16 pAb (#A304-192A, Bethyl Laboratories, Lucerne, Switzerland), rabbit anti-IGF2BP3 pAb (#14642-1-AP, Proteintech), or rabbit anti-IgG pAb (#30000-0-AP, Proteintech) was added into the lysates. Following incubation for 6 h at 4 °C, we isolated RNA from the precipitates to assess PFKM mRNA enrichment by quantitative PCR.
2.14Analysis of PFKM mRNA stabilitySi-METTL16-transfected or si-NC-introduced MHCC97 and Huh-7 cells were maintained in media containing 50 µg/mL Actinomycin D (Act D, Beyotime). We extracted RNA from treated cells to evaluate PFKM mRNA levels at 0, 3, 6 and 9 h thereafter.
2.15Chromatin immunoprecipitation (ChIP) assayTo validate the relationship of POU3F2 with the METTL16 promoter, we applied the ChIP Assay Kit (Beyotime) to perform ChIP experiments. Briefly, chromatin DNA was obtained from MHCC97 and Huh-7 cells and then subjected to fragmentation. Protein A/G Agarose and rabbit anti-POU3F2 pAb (#PA5-30124, Thermo Fisher Scientific) or rabbit anti-IgG pAb (#30000-0-AP, Proteintech) was mixed to generate bead-antibody complex, which was added into the suspension of DNA fragments. Following incubation for 8 h at 4 °C, we prepared RNA from the precipitates to detect the enrichment of the METTL16 promoter segments by quantitative PCR.
2.16Luciferase reporter assayWe generated METTL16 reporters by cloning the METTL16 promoter fragment harboring the predicted binding sequence (WT) or mutant target region (MUT) into the pGL3 basic vector (Thermo Fisher Scientific). MHCC97 and Huh-7 cells were co-transfected with WT-METTL16 or MUT-METTL16 and si-NC or si-POU3F2. The pRL-TK plasmid (Promega, Charbonnières, France) was also introduced into cells as the normalized control. At 48 h post transfection, we quantified the luciferase activity using Dual-Luciferase reporter system (Promega).
2.17Data analysisWe expressed data using mean±SD. For significance comparisons, we used Mann Whitney U test, unpaired t-test or ANOVA (one- or two-way). For survival analysis, we applied the Kaplan-Meier survival accompanied by log rank tests. Values of P < 0.05 were significant.
2.18Ethical statementHuman research protocols were authorized by the First Affiliated Hospital, Hengyang Medical School, University of South China Ethics and Scientific Committee. Mouse xenograft studies were conducted following protocols approved by the First Affiliated Hospital, Hengyang Medical School, University of South China Animal Care and Use Ethics Committee.
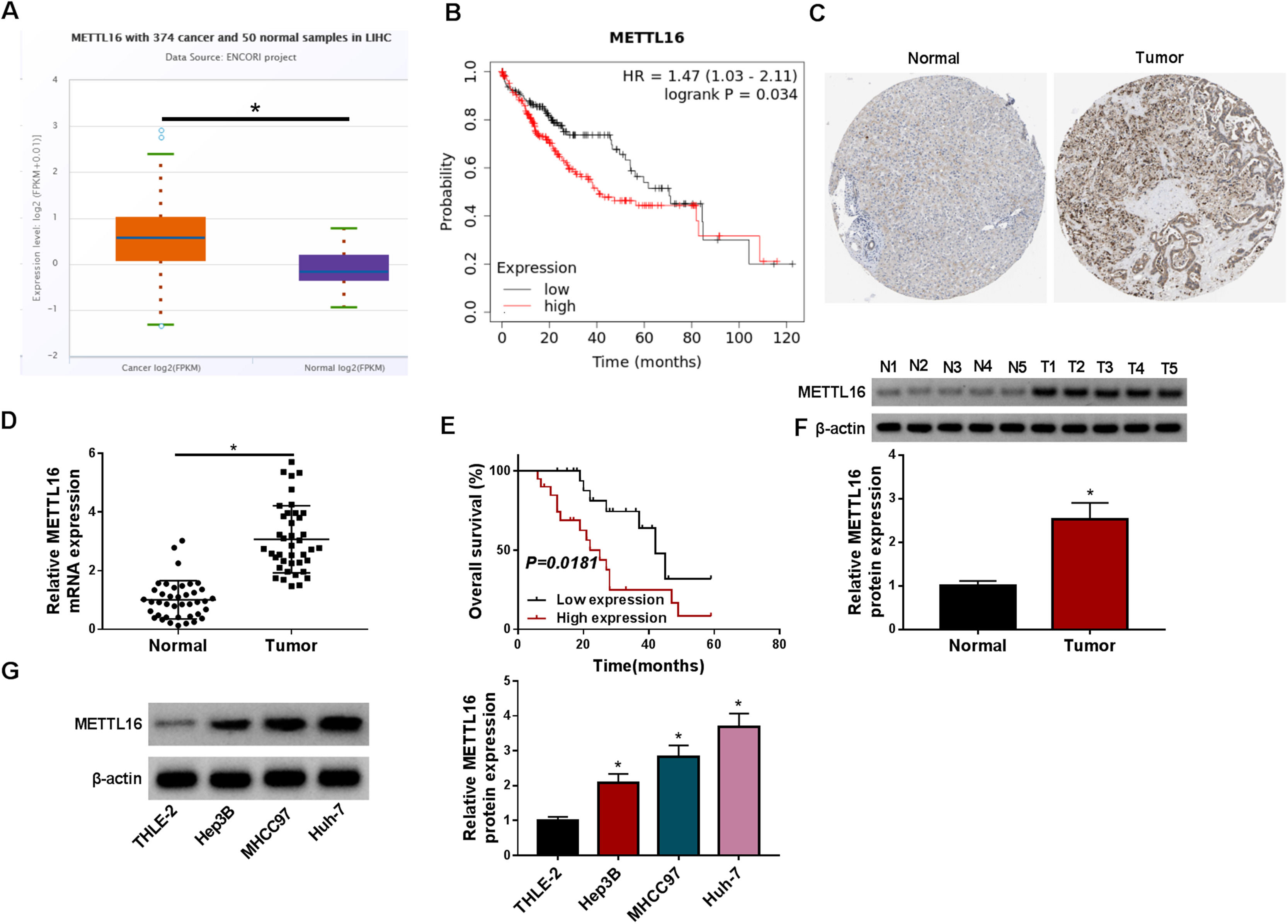
3Results3.1METTL16 is upregulated in HCC and its deficiency results in suppressed cell growth, invasiveness and sphere formationUsing the previously developed ENCORI algorithm, we observed the remarkably increased expression of METTL16 transcript in liver cancer (LIHC) compared with normal samples (Fig. 1A). We also used the online tool Kaplan-Meier Plotter to search for the association of METTL16 expression with HCC prognosis and found that high expression of METTL16 foreboded worse overall survival (Fig. 1B). The human protein atlas online resource showed the high expression of METTL16 protein in HCC specimens (Fig. 1C). Validating the online predictions, we collected a cohort of human clinical samples to obtain their RNA and protein extracts. Using quantitative PCR, we obtained evidence for elevated METTL16 mRNA expression in primary HCC tumors compared with matched healthy samples (Fig. 1D). The Kaplan-Meier survival curve revealed that high METTL16 expression (above median) predicted poor survival in HCC patients (Fig. 1E). Through immunoblotting, we also confirmed the upregulation of METTL16 protein in HCC tumors compared with adjacent non-cancerous tissues (Fig. 1F). Likewise, expression analysis of METTL16 in HCC cell lines showed elevated METTL16 protein levels in MHCC97, Huh-7, and Hep3B HCC cells relative to normal THLE-2 cells (Fig. 1G). Hence, increased expression of METTL16 observed in HCC is tightly linked to HCC prognosis.
METTL16 expression is increased in HCC, and elevated METTL16 expression predicts poor HCC prognosis. (A) ENCORI algorithm revealing the upregulation of METTL16 transcript in liver cancer (LIHC) compared with normal samples. (B) Kaplan-Meier Plotter database showing the association of METTL16 expression with HCC prognosis. (C) The human protein atlas resource unveiling the high expression of METTL16 protein in HCC specimens. (D) Validation of high METTL16 transcript in clinical HCC tumors (n = 40) compared with matched healthy samples (n = 40). P-value based on the Mann-Whitney U test. (E) Kaplan-Meier survival curve revealing the positive correlation of high METTL16 expression (above median) with poor survival. P-value based on the Log-rank (Mantel-Cox) test. (F) Validation of high METTL16 protein levels in clinical HCC tumors (n = 5) compared with matched healthy samples (n = 5). P-value based on a two-tailed Student's t-test. (G) Expression of METTL16 protein in MHCC97, Huh-7, and Hep3B HCC cells and non-tumor THLE-2 cells, as evaluated by immunoblotting (n = 3). P based on one-way ANOVA followed by Tukey-Kramer post hoc test. *P < 0.05.
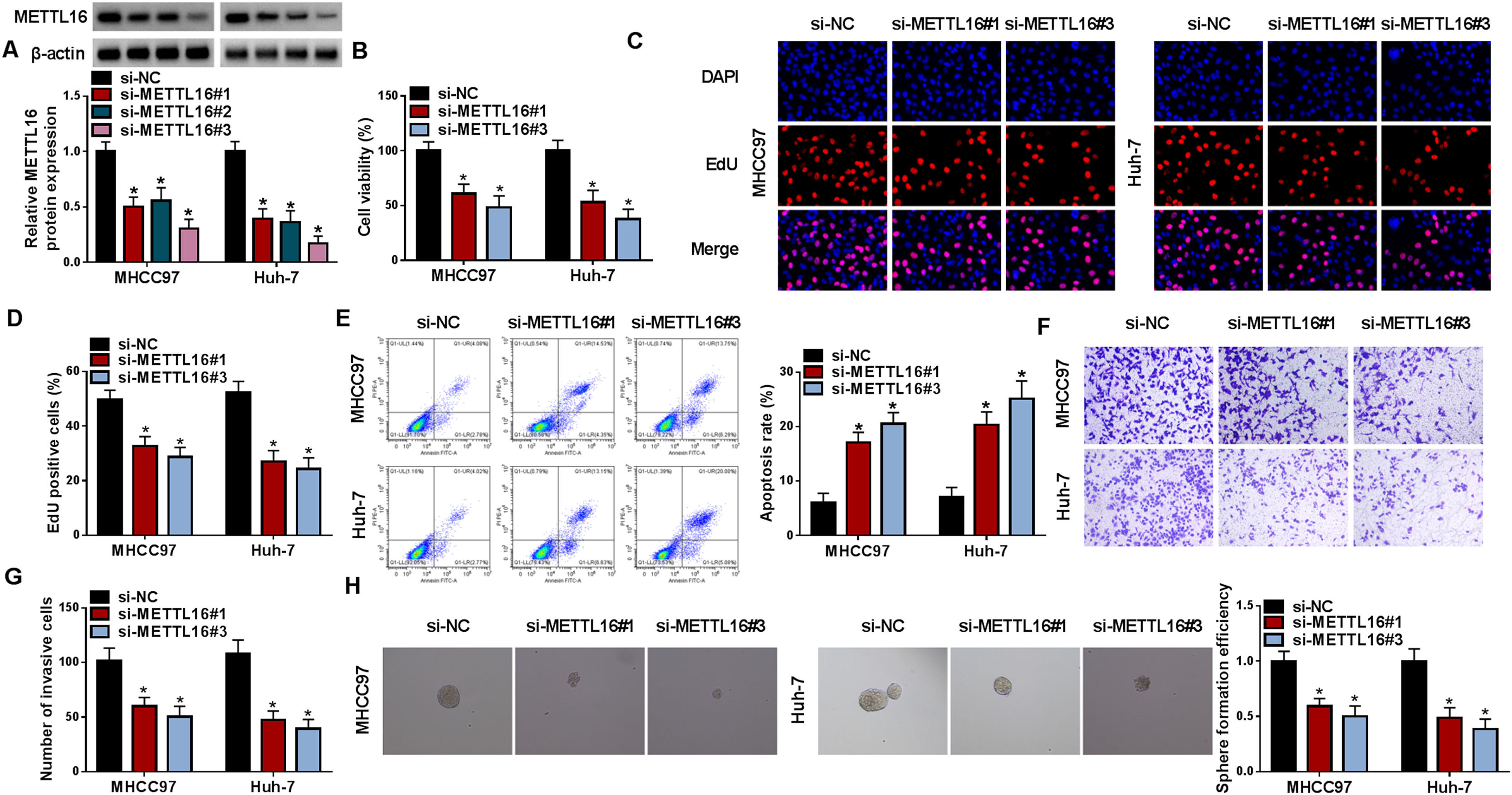
To examine whether upregulation of METTL16 participates in the carcinogenesis of HCC, we sought to reduce its expression to observe the consequences in MHCC97 and Huh-7 HCC cells, which exhibited more significantly upregulated METTL16 (Fig. 1G). For that purpose, three METTL16 siRNAs (si-METTL16#1, si-METTL16#2 or si-METTL16#3) were transfected separately into the two HCC cell lines, and their knockdown efficiencies were validated by immunoblotting (Fig. 2A). Since si-METTL16#1 and si-METTL16#3 had a stronger knockdown efficacy than si-METTL16#2 (Fig. 2A), the two siRNAs were used in the subsequent experiments. After METTL16 depletion, we observed a significant decrease in cell viability and the number of EdU positive cells (Fig. 2B–2D). Conversely, the apoptosis ratio was remarkably higher in METTL16-depleted HCC cells than that in si-NC controls (Fig. 2E). Using transwell assay, we were able to show that METTL16-depleted HCC cells displayed suppressed invasiveness (Fig. 2F and 2G). Furthermore, the efficiency of sphere formation of MHCC97 and Huh-7 HCC cells was markedly diminished following METTL16 deficiency (Fig. 2H). Taken together, our results suggest that upregulated METTL16 plays a promoting role in HCC tumorigenesis.
METTL16 deficiency suppresses HCC cell growth, invasiveness and sphere formation in vitro. (A) Expression of METTL16 protein by immunoblotting in MHCC97 and Huh-7 HCC cells transfected with si-NC, si-METTL16#1, si-METTL16#2 or si-METTL16#3. (B-H) MHCC97 and Huh-7 HCC cells were introduced with si-NC, si-METTL16#1, or si-METTL16#3. (B) CCK-8 cell viability assay in transfected MHCC97 and Huh-7 HCC cells. (C and D) EdU cell proliferation assay with MHCC97 and Huh-7 HCC cells transfected as indicated. (E) Flow cytometry for cell apoptosis in MHCC97 and Huh-7 HCC cells after the indicated transfection. (F and G) Transwell invasion assay with thansfected MHCC97 and Huh-7 HCC cells. (H) Sphere formation of MHCC97 and Huh-7 HCC cells after the indicated transfection was assessed. n = 3 for A-H. *P < 0.05 based on two-way ANOVA followed by Sidak's multiple comparisons test.
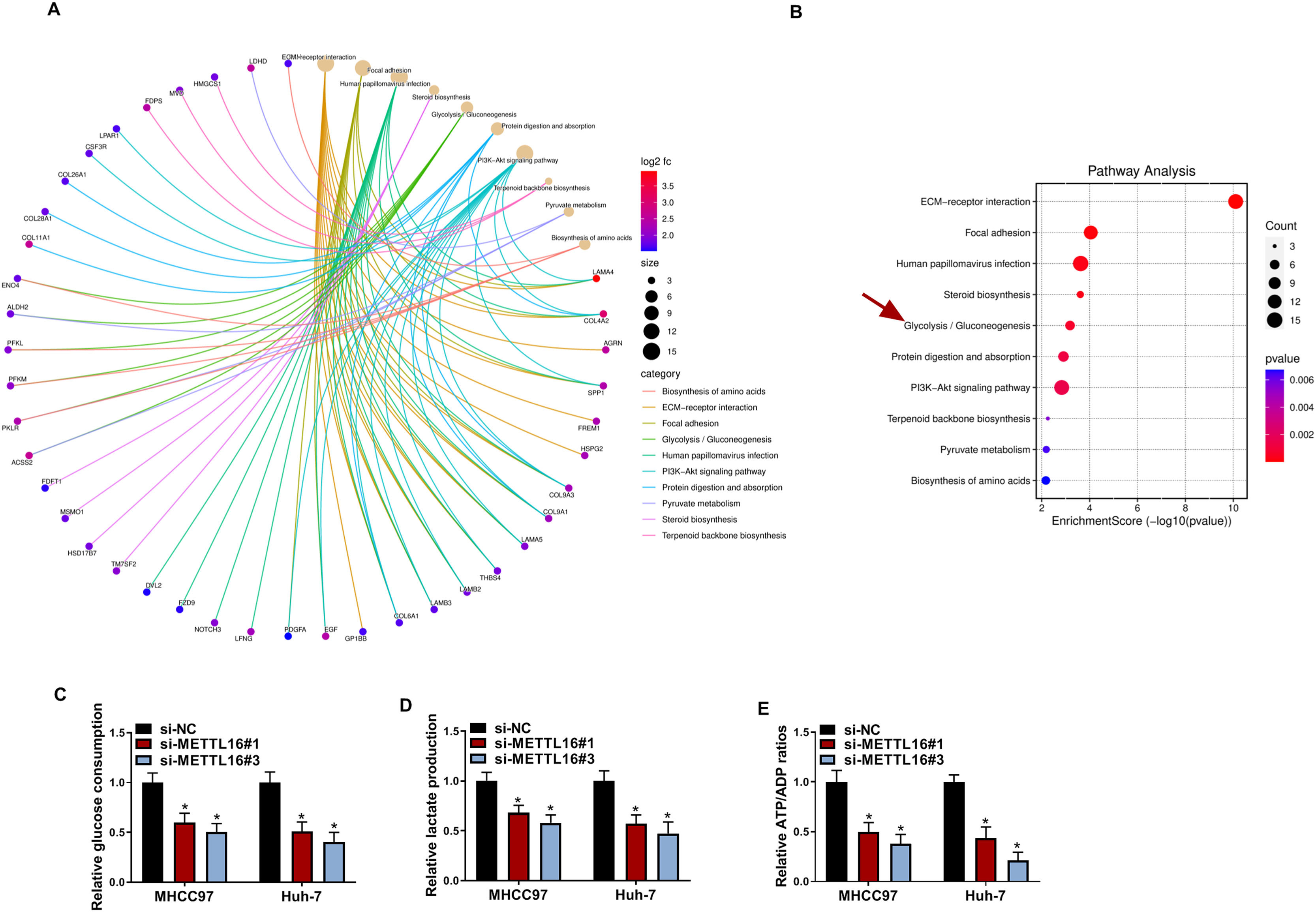
In an attempt to evaluate the mechanism behind the action of METTL16 in HCC, we performed a bioinformatic analysis for the downstream targets of METTL16. The GSE224008 dataset revealed many differentially expressed genes (DEGs) after METTL16 silencing in Huh-7 liver cancer cells. Through KEGG pathway enrichment analysis, we found that these DEGs were closely related to various important pathways, including glycolysis/gluconeogenesis pathway (Fig. 3A and 3B). We thus hypothesized that METTL16 might be involved in HCC glycolysis. By applying commercially available assay kits, we found significantly decreased glucose consumption, lactate production, and ATP/ADP ratio in METTL16-silenced MHCC97 and Huh-7 HCC cells compared with si-NC control cells (Fig. 3C–3E). These data imply that upregulated METTL16 may contribute to enhanced glycolysis in HCC.
METTL16 depletion inhibits HCC cell glycolysis in vitro. (A and B) GSE224008 dataset revealed many DEGs after METTL16 silencing in Huh-7 liver cancer cells. KEGG pathway enrichment analysis of these DEGs was performed. (C-E) Cell glucose consumption level (C), lactate production level (D), and ATP/ADP ratio (E) in MHCC97 and Huh-7 HCC cells transfected with si-NC, si-METTL16#1, or si-METTL16#3 were evaluated using corresponding assay kits (n = 3). P based on two-way ANOVA followed by Sidak's multiple comparisons test. *P < 0.05.
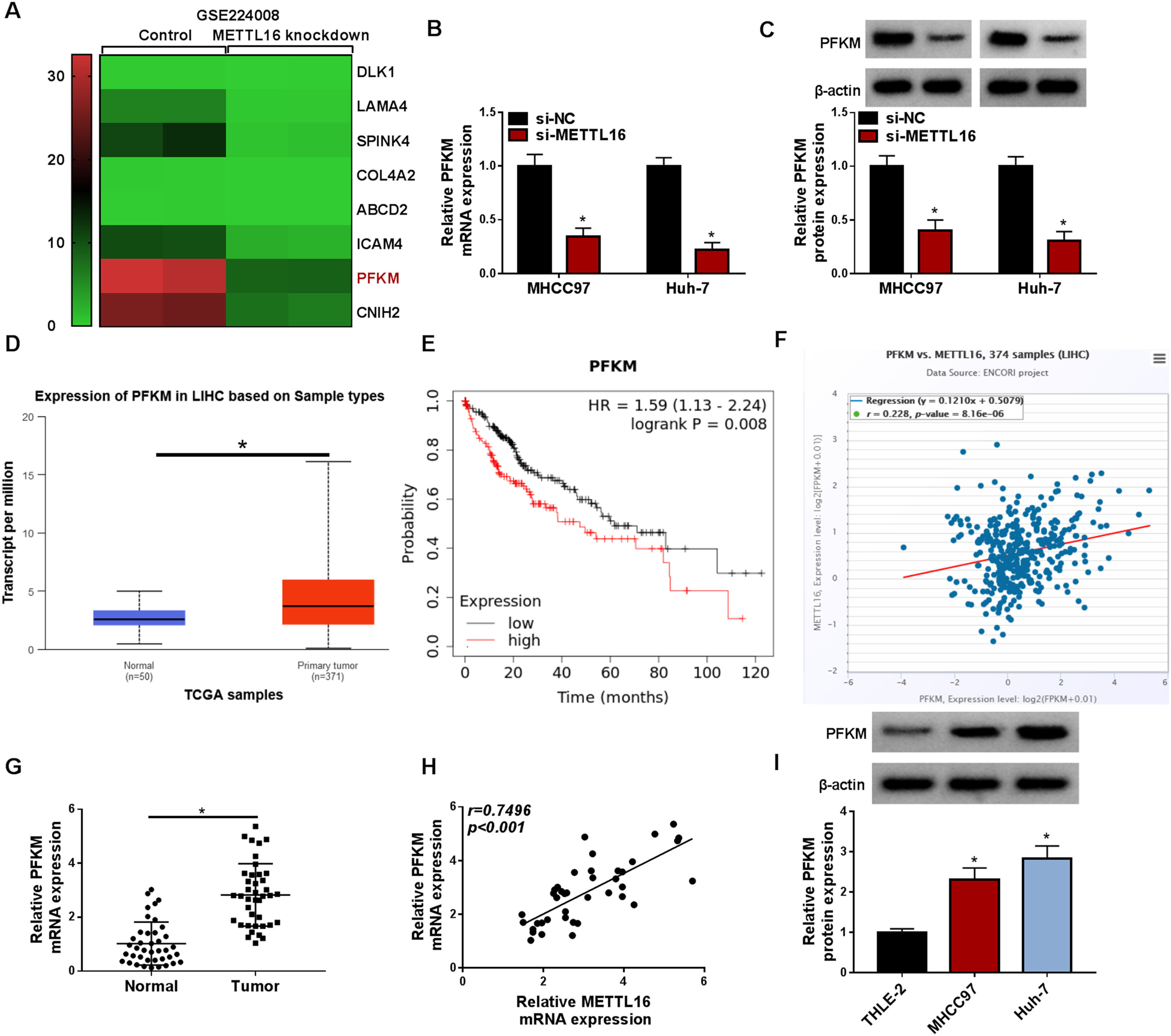
After studying the modulation of METTL16 in HCC glycolysis, we next investigated the molecular basis underlying this modulation. Further observation of the GSE224008 dataset showed that PFKM, a crucial rate-limiting enzyme in glycolysis that has been identified as a potential driver in HCC [13], was strongly reduced in METTL16-depleted Huh-7 cells (Fig. 4A). Using mRNA and protein expression analyses, we were able to demonstrate that METTL16 knockdown led to a significant decrease in PFKM expression in MHCC97 and Huh-7 HCC cells (Fig. 4B and 4C). The open-access TCGA database predicted the elevated expression of PFKM transcript in liver cancer (Fig. 4D). Elevated PFKM levels foreboded worse survival in HCC, examined by the Kaplan-Meier Plotter web (Fig. 4E). In addition, the ENCORI algorithm revealed the possible positive expression correlation of PFKM with METTL16 in liver cancer (Fig. 4F). Using quantitative PCR, we also obtained evidence for the increased expression of PFKM mRNA transcript and the positive expression correlation between PFKM and METTL16 in primary HCC tumors (Fig. 4G and 4H). Additionally, using immunoblotting, we observed increased levels of PFKM protein in MHCC97 and Huh-7 HCC cells compared with non-tumor THLE-2 cells (Fig. 4I).
METTL16 affects PFKM expression in HCC cells. (A) GSE224008 dataset showing the decrease of PFKM transcript in Huh-7 liver cancer cells with METTL16 knockdown. (B) Expression of PFKM mRNA transcript in MHCC97 and Huh-7 HCC cells transfected with si-NC or si-METTL16 (the 1:1 ratio mix of si-METTL16#1:si-METTL16#3) by quantitative PCR (n = 3). P based on two-way ANOVA followed by Sidak's multiple comparisons test. (C) Expression of PFKM protein in si-NC- or si-METTL16-transfected HCC cells (n = 3). P based on two-way ANOVA followed by Sidak's multiple comparisons test. (D) TCGA database showing the high expression of PFKM mRNA in liver cancer (LIHC) tissues compared with normal samples. (E) Kaplan-Meier Plotter database predicting the association of PFKM expression with HCC prognosis. (F) ENCORI algorithm predicting the positive expression correlation of PFKM with METTL16. (G) Validation of high PFKM mRNA transcript in clinical HCC tumors (n = 40) compared with matched healthy samples (n = 40). P-value based on the Mann-Whitney U test. (H) Validation of the positive expression correlation of PFKM with METTL16 in clinical HCC tumors (n = 40) by Pearson's correlation analysis. (I) Expression of PFKM protein in MHCC97, Huh-7, and Hep3B HCC cells and non-tumor THLE-2 cells by immunoblotting (n = 3). P based on one-way ANOVA followed by Tukey-Kramer post hoc test. *P < 0.05.
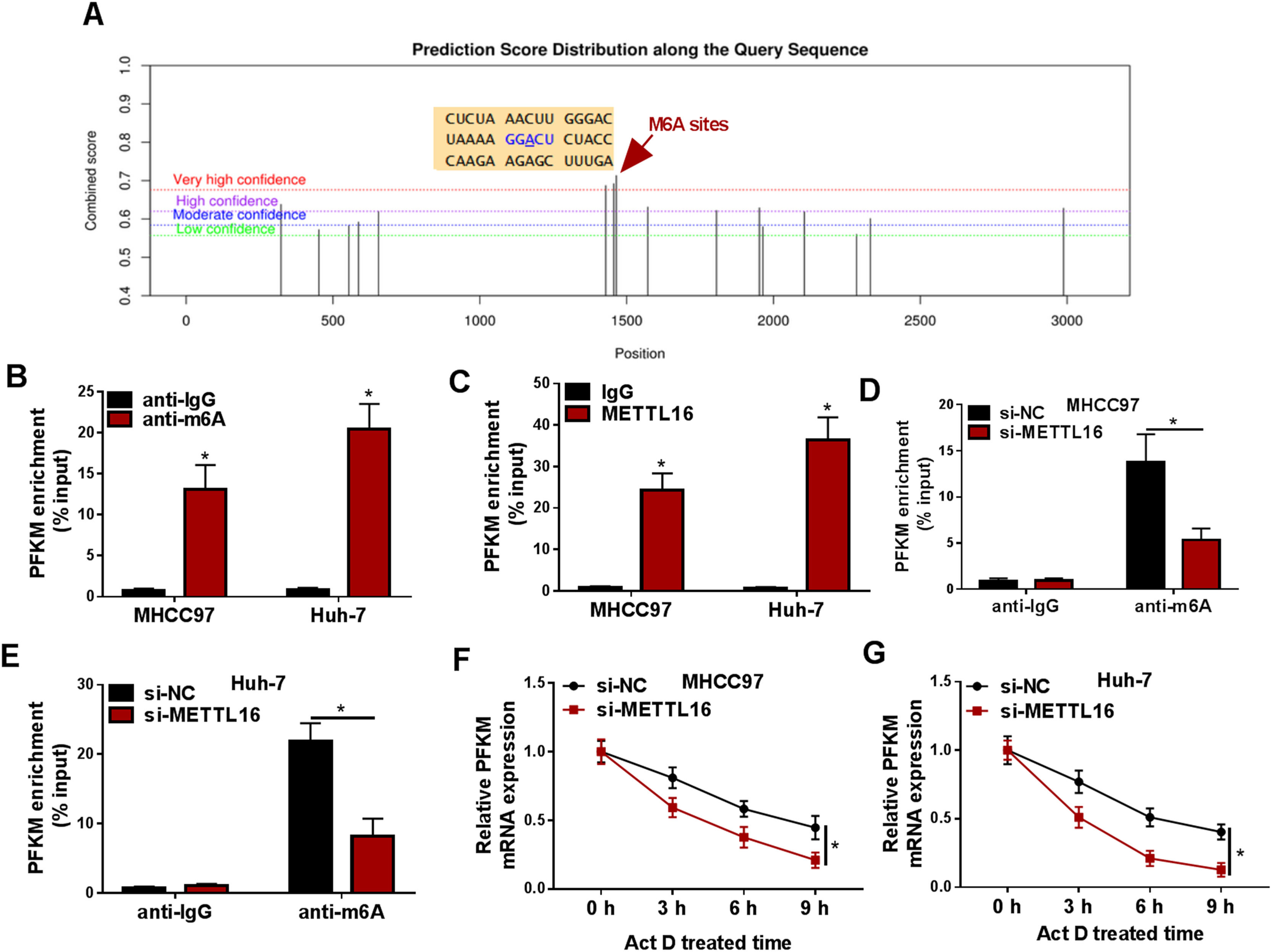
Dysregulation of METTL16, an atypical methyltransferase, has been reported to play a pro-tumorigenic role via an m6A-dependent mechanism [7,14]. Thus, we further studied whether METTL16 affects PFKM expression via the m6A-dependent mechanism. The online website SRAMP predicted the putative m6A methylation sites within PFKM mRNA (Fig. 5A). MeRIP experiments using an anti-m6A antibody validated the existence of m6A-modified PFKM mRNA in the two HCC cell lines, as presented by the enrichment of PFKM mRNA transcript in m6A-associating precipitates (Fig. 5B). RIP experiments using an anti-METTL16 antibody also confirmed the direct relationship of METTL16 with PFKM mRNA (Fig. 5C). Furthermore, si-METTL16-mediated METTL16 depletion significantly reduced the levels of m6A-modified PFKM mRNA in MHCC97 and Huh-7 cells (Fig. 5D and 5E). Interestingly, using Act D treatment to inhibit transcription, we observed that silencing of METTL16 strongly weakened the stability of PFKM mRNA in both HCC cell lines (Fig. 5F and 5G). All these observations demonstrate that METTL16 mediates m6A methylation of PFKM mRNA to stability this mRNA transcript.
METTL16 mediates m6A methylation to stabilize PFKM mRNA. (A) SRAMP website predicted the putative m6A methylation sites within PFKM mRNA. (B and C) MeRIP with anti-m6A antibody (B) and RIP with anti-METTL16 antibody (C) were performed using lysates of MHCC97 and Huh-7 HCC cells. Enrichment of PFKM transcript was gauged by quantitative PCR (n = 3). (D and E) MeRIP with anti-m6A antibody was done using lysates of si-NC- or si-METTL16-transfected cells, and followed by detection of PFKM transcript enrichment by quantitative PCR (n = 3). (F and G) Si-NC- or si-METTL16-transfected MHCC97 and Huh-7 cells were subjected to Act D treatment for the indicated time and checked for PFKM mRNA levels by quantitative PCR (n = 3). *P < 0.05 based on two-way ANOVA followed by Sidak's multiple comparisons test.
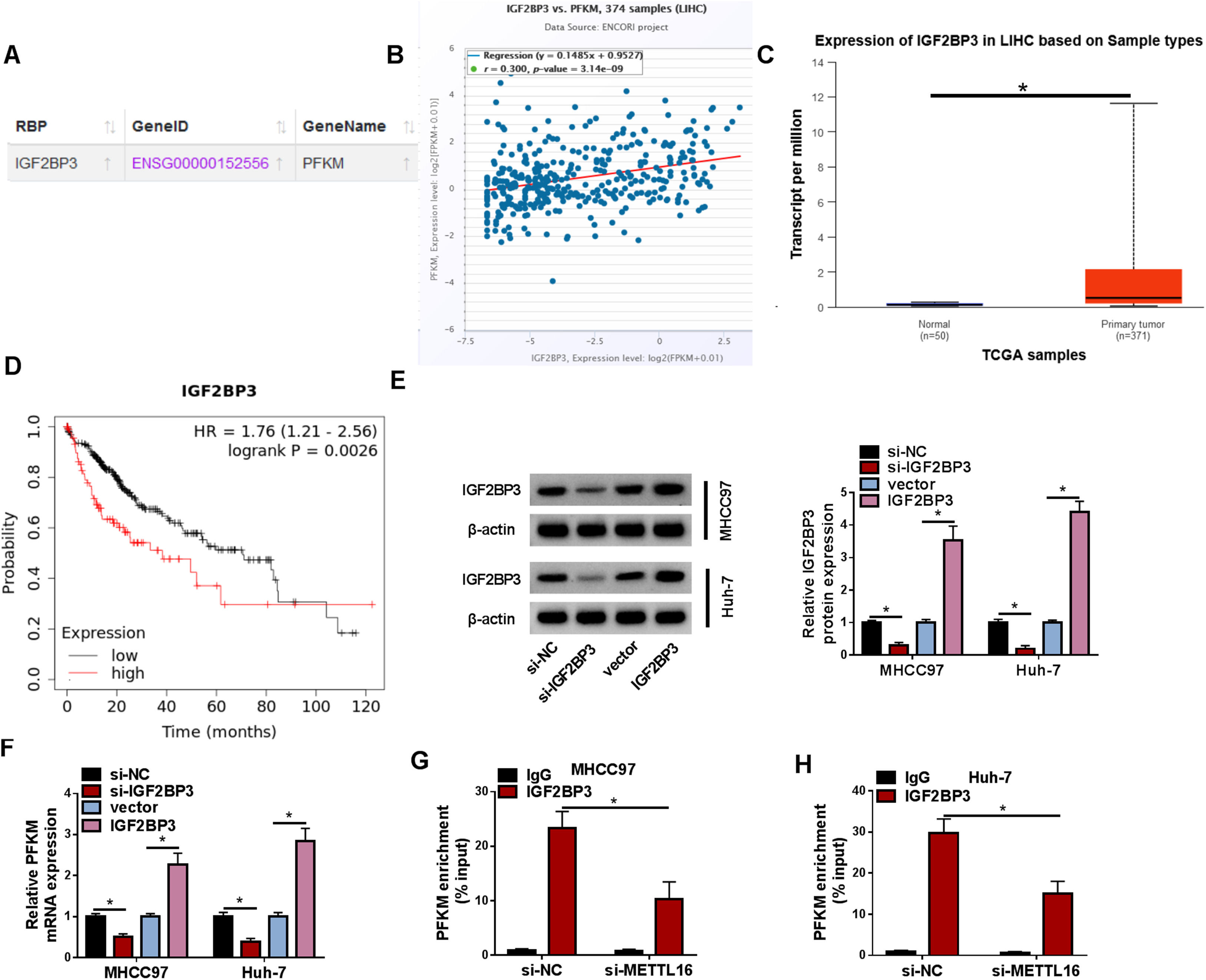
The knowledge of RNA m6A modification has revealed essential roles of m6A binding proteins (m6A readers) in RNA stability [15]. Therefore, we also performed bioinformatic and experimental analyses to identify an m6A reader associating with the regulation of METTL16 in PFKM mRNA stability. Using the online ENCORI algorithm, we observed the putative binding of IGF2BP3, a crucial m6A reader that is reported to stabilize target mRNAs in HCC [16,17], to PFKM mRNA (Fig. 6A). The ENCORI algorithm also predicted the positive expression correlation between PFKM and IGF2BP3 in liver cancer (Fig. 6B). The TCGA database showed the upregulation of IGF2BP3 transcript in liver cancer samples (Fig. 6C), and the Kaplan-Meier Plotter tool revealed that IGF2BP3 upregulation predicted poor prognosis in this disease (Fig. 6D). The transfection efficiencies of si-IGF2BP3 and IGF2BP3 expression construct were validated by immunoblotting (Fig. 6E). Through quantitative PCR, we demonstrated the positive modulation of IGF2BP3 in PFKM mRNA expression in MHCC97 and Huh-7 cells (Fig. 6F). Interestingly, METTL16 depletion significantly diminished the relationship of IGF2BP3 with PFKM mRNA, as evidenced by the reduction of PFKM mRNA enrichment levels in IGF2BP3-associating precipitates (Fig. 6G and 6H). Taking together, we conclude that METTL16 enhances PFKM mRNA stability via an IGF2BP3-m6A-dependent manner.
IGF2BP3 influences PFKM mRNA expression. (A) ENCORI algorithm observed the putative binding of IGF2BP3 to PFKM mRNA transcript. (B) ENCORI algorithm predicted the positive expression correlation between PFKM and IGF2BP3 in liver cancer (LIHC). (C) TCGA database showed the upregulation of IGF2BP3 transcript in LIHC samples. (D) Kaplan-Meier Plotter tool revealed that IGF2BP3 upregulation predicted poor prognosis in LIHC patients. (E) Expression of IGF2BP3 protein in MHCC97 and Huh-7 cells transfected with si-NC, si-IGF2BP3, vector, or IGF2BP3 expression construct (n = 3). P based on two-way ANOVA followed by Sidak's multiple comparisons test. (F) Expression of PFKM mRNA in MHCC97 and Huh-7 cells transfected as indicated (n = 3). P based on two-way ANOVA followed by Sidak's multiple comparisons test. (G and H) RIP with anti-IGF2BP3 antibody was done using lysates of si-NC- or si-METTL16-transfected cells, followed by detection of PFKM transcript enrichment by quantitative PCR (n = 3). P based on two-way ANOVA followed by Sidak's multiple comparisons test. *P < 0.05.
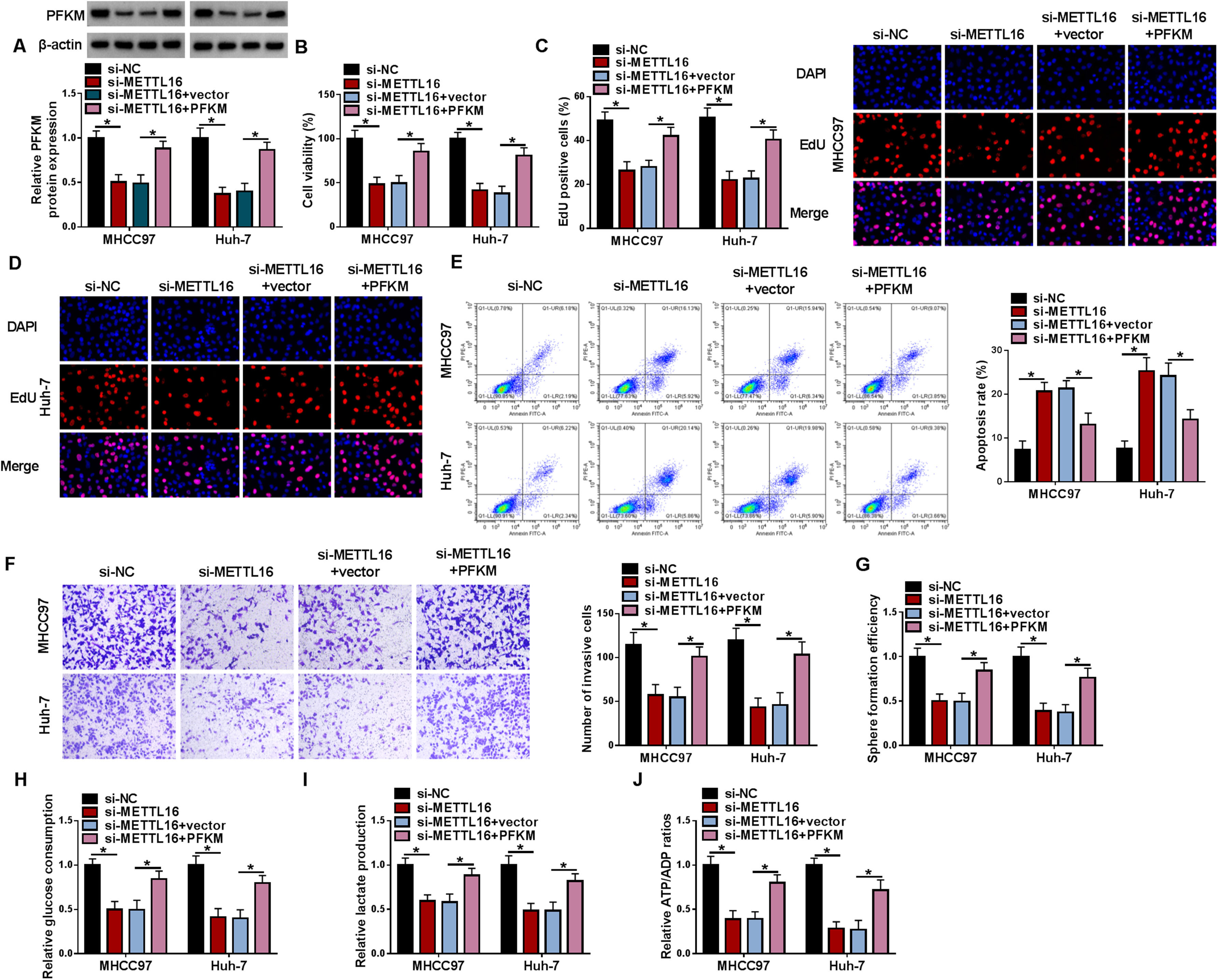
Our findings have established the positive regulation of METTL16 in PFKM mRNA stability in HCC cells. Accordingly, we further performed a series of in vitro and in vivo rescue assays to elucidate whether METTL16 affects HCC carcinogenesis through PFKM. In METTL16-silenced MHCC97 and Huh-7 cells, a PFKM construct was used to elevate PFKM expression (Fig. 7A). Restored PFKM levels significantly reversed METTL16 depletion-mediated cell in vitro viability inhibition (Fig. 7B), proliferation suppression (Fig. 7C and 7D), apoptosis promotion (Fig. 7E), invasion repression (Fig. 7F), and sphere formation reduction (Fig. 7G) in MHCC97 and Huh-7 cells. Likewise, PFKM increase strongly counteracted si-METTL16-imposed in vitro diminishment of glucose consumption (Fig. 7H), lactate production (Fig. 7I), and ATP/ADP ratio (Fig. 7J) of MHCC97 and Huh-7 cells.
Restored PFKM expression reverses METTL16 deficiency-mediated cell phenotype alterations in vitro. (A-J) MHCC97 and Huh-7 HCC cells were subjected to transfection of si-NC, si-METTL16, si-METTL16+vector, or si-METTL16+PFKM expression construct. (A) Expression of PFKM protein by immunoblotting in transfected cells. (B) CCK-8 cell viability assay in transfected MHCC97 and Huh-7 HCC cells. (C and D) EdU cell proliferation assay with MHCC97 and Huh-7 HCC cells transfected as indicated. (E) Flow cytometry for cell apoptosis in MHCC97 and Huh-7 HCC cells after the indicated transfection. (F) Transwell invasion assay with thansfected MHCC97 and Huh-7 HCC cells. (G) Sphere formation of MHCC97 and Huh-7 HCC cells after the indicated transfection was assessed. (H-J) Cell glucose consumption level, lactate production level, and ATP/ADP ratio in MHCC97 and Huh-7 HCC cells transfected as indicated using corresponding assay kits. n = 3 for A-J. *P < 0.05 based on two-way ANOVA followed by Sidak's multiple comparisons test.
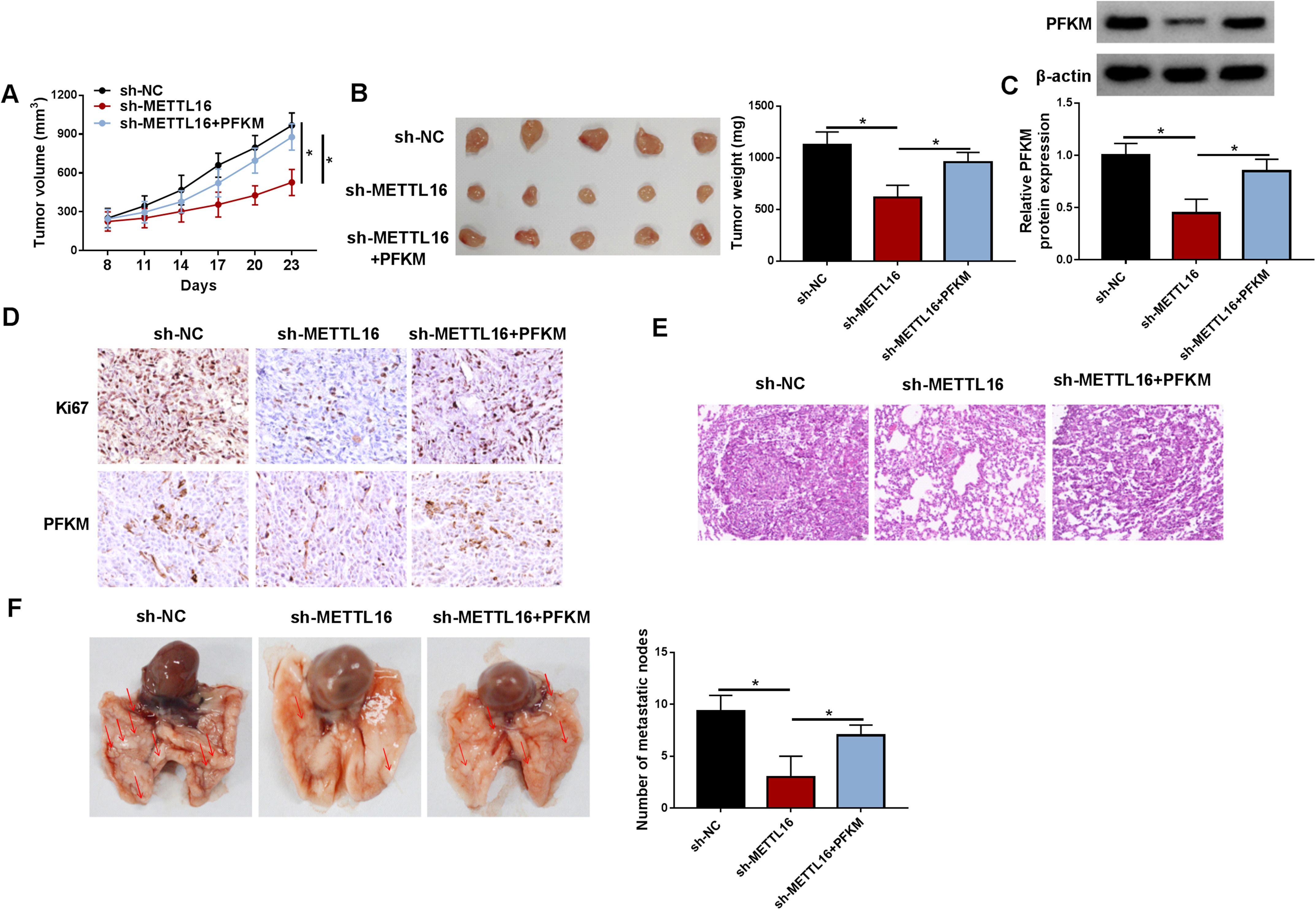
We also performed in vivo experiments by generating MHCC97 mouse xenograft tumors accompanied with or without intratumoral administration of the PFKM expression construct. Sh-METTL16-injected mice produced significantly smaller tumors than sh-NC controls (Fig. 8A and 8B). Whereas, administration of the PFKM construct strongly abolished this phenomenon (Fig. 8A and 8B). Sh-METTL16 MHCC97 xenografts exhibited reduced PFKM protein expression, while the PFKM construct reversed the reduction (Fig. 8C and 8D). In addition, xenografts formed by sh-METTL16-infected MHCC97 cells had fewer cells stained for Ki67, and elevated PFKM expression enhanced the number of Ki67 positive tumor cells (Fig. 8D). We then asked whether the METTL16/PFKM axis would result in distant metastasis. At week 6 after cell implantation, H&E staining revealed the occurrence of MHCC97 tumor lung metastasis in sh-NC mice (Fig. 8E). The number of metastatic nodes also supported the occurrence of tumor lung metastasis (Fig. 8F). Sh-METTL16 MHCC97 xenografts displayed decreased lung metastasis, whereas elevated PFKM expression strongly enhanced MHCC97 tumor lung metastasis (Fig. 8E and 8F). Collectively, we establish the notion that PFKM works as a functional effector of METTL16 in promoting HCC.
Restored PFKM expression abolishes METTL16 deficiency-driven tumor growth suppression in vivo. Sh-NC- or sh-METTL16 lentivirus-infected MHCC97 cells were implanted subcutaneously or intravenously into nude mice, with or without administration of the PFKM expression lentivirus. n = 8 for each group. Five mice were used for xenograft collection at day 23, and three mice were used for lung collection at week 6. (A) Growth curves of MHCC97 xenografts. (B) Images and mean weight of MHCC97 xenografts on day 23. (C) Expression of PFKM protein by immunoblotting in MHCC97 xenografts. (D) Expression of PFKM and Ki67 by immunohistochemistry in MHCC97 xenografts. (E) H&E staining of lung tissues of xenograft mice. (F) The number of metastasis nodes in lung tissues of xenograft mice. *P < 0.05 based on one-way ANOVA followed by Tukey-Kramer post hoc test.
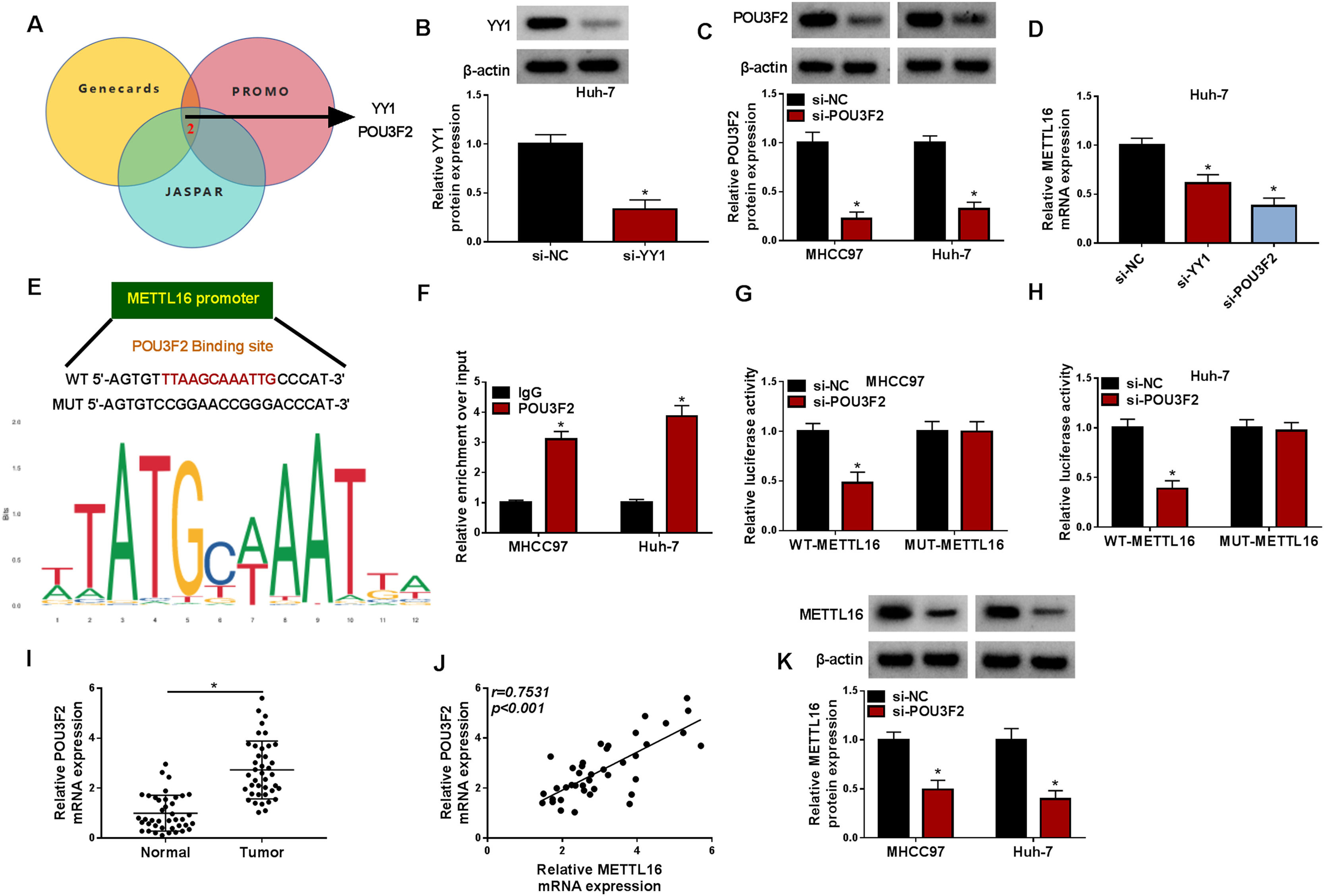
Finally, we also investigated the mechanism underlying METTL16 upregulation in HCC. Dysregulation of transcription factors (TFs) is directly associated with liver tumorigenesis [18]. We therefore decided to elucidate the upstream TFs in promoting METTL16 transcription using three TF-predicting algorithms (Genecards, PROMO and JASPAR). A total of two putative TFs (YY1 and POU3F2) that overlapped among these algorithms were found (Fig. 9A). The depletion efficacy of si-YY1 or si-POU3F2 was confirmed by immunoblotting in HCC cells (Fig. 9B and 9C). Interestingly, the expression of METTL16 mRNA transcript was markedly decreased after depletion of YY1 or POU3F2 in Huh-7 cells (Fig. 9D). Because of the more significantly reduction by si-POU3F2 (Fig. 9D), we focused on POU3F2 in the present work. The JASPAR algorithm predicted the putative binding sites (TTAAGCAAATTG) for POU3F2 in the METTL16 promoter (Fig. 9E). ChIP experiments with an anti-POU3F2 antibody validated the relationship of POU3F2 with the METTL16 promoter (Fig. 9F). To examine the modulation of POU3F2 in METTL16 transcription via the target region, we generated METTL16 promoter reporter constructs containing the putative binding sequence (WT-METTL16) or mutant target region (MUT-METTL16). Luciferase assays showed that si-POU3F2-driven POU3F2 depletion strongly reduced the luciferase activity of WT-METTL16, but not MUT-METTL16 (Fig. 9G and 9H). In addition, in clinical HCC samples, POU3F2 expression was enhanced and positively related to METTL16 expression (Fig. 9I and 9J). Further observation of the influence of POU3F2 in METTL16 expression confirmed that POU3F2 silencing led to a significant reduction in METTL16 protein expression (Fig. 9K). These results demonstrate that upregulated POU3F2 is a contributor to METTL16 increase by enhancing METTL16 transcription in HCC.
POU3F2 enhances METTL16 transcription in HCC cells. (A) Genecards, PROMO and JASPAR algorithms predicted the putative TFs in regulating METTL16 transcription. (B and C) The transfection efficiencies of si-YY1 or si-POU3F2 were confirmed by immunoblotting. P-value based on a two-tailed Student's t-test or two-way ANOVA followed by Sidak's multiple comparisons test. (D) Expression of METTL16 transcript in Huh-7 cells after depletion of YY1 or POU3F2. P based on one-way ANOVA followed by Tukey-Kramer post hoc test. (E) JASPAR algorithm predicted the putative binding sites between POU3F2 and the METTL16 promoter. (F) ChIP with an anti-POU3F2 antibody was done using lysates of MHCC97 and Huh-7 cells. P based on two-way ANOVA followed by Sidak's multiple comparisons test. (G and H) Luciferase assays in cells transfected with si-POU3F2 or si-NC and WT-METTL16 or MUT-METTL16. P based on two-way ANOVA followed by Sidak's multiple comparisons test. (I) Validation of high POU3F2 mRNA transcript in clinical HCC tumors (n = 40) compared with matched healthy samples (n = 40). P-value based on the Mann-Whitney U test. (J) Validation of the positive expression correlation of POU3F2 with METTL16 in clinical HCC tumors (n = 40) by Pearson's correlation analysis. (K) Expression of METTL16 protein in cells after transfection with si-POU3F2 or si-NC. P based on two-way ANOVA followed by Sidak's multiple comparisons test. *P < 0.05.
Advanced therapies are beneficial to early stage HCC and fail to provide great benefit for HCC at the advanced stage. Thus, investigating the molecular pathogenesis of HCC is crucial for developing innovative therapeutic approaches against HCC. Being the second methyltransferase identified, m6A writer METTL16 is aberrantly expressed in various cancers, and its deregulation is strongly linked to human carcinogenesis [19]. Recent work has documented the pro-tumorigenic role of METTL16 in HCC through an m6A-dependent mode [9,10,20]. Consistently, our findings provide support to the fact that increased METTL16 levels observed in HCC are tightly related to HCC prognosis, and METTL16 plays a promoting role in HCC tumorigenesis. Increased glycolysis in cancer cells contributes to oncogenesis and cancer progression. Modulation of glycolysis has been proposed for improving cancer treatment [4]. Our results also show that depletion of METTL16 results in suppressed glycolysis in HCC cells. Hence, METTL16 inhibitors may offer the potential opportunity to combat HCC.
As a crucial regulatory enzyme in glycolysis, PFKM is capable of enhancing glycolysis and thus exerts a promoting impact on human tumorigenesis [21]. An important example is that mutation of PFKM has been described to hinder cancer cell proliferation, xenograft growth and metastasis in ovarian cancer [22]. YTHDC1-mediated PFKM mRNA m6A methylation takes part in osteosarcoma pathogenesis by modulating glycolysis [23]. Furthermore, PFKM is implicated in HCC cell growth and glycolysis [24]. Elevated expression of PFKM contributes to enhanced glycolysis, growth, and invasiveness in HCC cells [13]. These reports suggest the oncogenic effect of PFKM on human malignancies, including HCC. Here, we have found the positive modulation of METTL16 in PFKM expression. Moreover, METTL16 can mediate m6A methylation to stabilize PFKM mRNA. IGF2BP3, a key m6A reader, functions as a critical player in HCC by upgrading target mRNA stability [16,17]. Importantly, we demonstrate, for the first time, that METTL16-mediated PFKM mRNA stabilization is IGF2BP3-dependent in HCC cells. Through in vitro and in vivo rescue assays, we define that PFKM has the ability to work as a functional effector of METTL16 in promoting HCC.
Epigenetic deregulation plays a pro-tumorigenic role that participates in liver carcinogenesis [25]. In this paper, we further highlight an epigenetic event driving METTL16 upregulation in HCC. Considering the crucial functions of TFs in gene expression, we uncover that TF POU3F2 promotes the transcription of METTL16 in HCC cells. POU3F2 has been identified that operates as a potent oncogenic factor in many types of human malignancies, such as breast cancer, prostate cancer and glioblastoma [26–28]. Moreover, the promoting role of POU3F2 in HCC has been established by previously reported literature [29]. Hence, it appears that POU3F2 upregulates METTL16 to promote HCC development. Nonetheless, additional studies are needed to further explain the details of the cascade in promoting HCC progression.
With these observations, the si-METTL16 or sh-METTL16 construct appears to represent a highly promising candidate for anti-HCC therapy. It exhibits a dual mechanism of action, not only inhibiting the growth and invasive capabilities of HCC cells but also effectively blocking their glycolysis potential. We envision that METTL16 inhibitors could serve as a pivotal starting point for the development of molecularly targeted therapies against HCC. More studies are required to assess the safety, long-term effectiveness, and potential of such agents in diverse experimental models.
5ConclusionsIn conclusion, we provide novel evidence for the pro-tumorigenic role of METTL16 in HCC. We propose METTL16-mediated PFKM mRNA stability via IGF2BP3 as a mechanism by which METTL16 promotes HCC progression, and we further define that TF POU3F2 drives METTL16 upregulation. Our findings highlight the implication of the POU3F2/METTL16/PFKM axis in HCC pathogenesis. Targeting the novel axis represents the potential opportunity to combat HCC.
FundingThis work was supported by Hunan Provincial Health Commission guiding project (No.D202303039177).
Author contributionsAll authors contributed significantly to this work. Their contributions include the conceptualization of the study, experimental design, data collection, and analysis. The authors also participated in the drafting and critical revision of the manuscript for important intellectual content. Additionally, all authors have reviewed and approved the final version of the manuscript and agree to be accountable for all aspects of the work, ensuring its accuracy and integrity.