During the last 30 years there has been a dissemination of plasmid-mediated β-lactamases in Enterobacteriaceae in Brazil. Extended spectrum β-lactamases (ESBL) are widely disseminated in the hospital setting and are detected in a lower frequency in the community setting. Cefotaximases are the most frequently detected ESBL type and Klebsiella pneumoniae is the predominant species among ESBL producers. Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae became widely disseminated in Brazil during the last decade and KPC production is currently the most frequent resistance mechanism (96.2%) in carbapenem resistant K. pneumoniae. To date KPC-2 is the only variant reported in Brazil. Polymyxin B resistance in KPC-2-producing K. pneumoniae has come to an alarming rate of 27.1% in 2015 in São Paulo, the largest city in Brazil. New Delhi metallo-β-lactamase was detected in Brazil in 2013, has been reported in different Brazilian states but are not widely disseminated. Antimicrobial resistance in Enterobacteriaceae in Brazil is a very serious problem that needs urgent actions which includes both more strict adherence to infection control measures and more judicious use of antimicrobials.
The first reports on antimicrobial resistance in Gram-negative rods from Brazil, available at PubMed, were restricted to community-acquired infections. These reports were on sulfadiazine resistance in Escherichia coli, Shigella and Salmonella and dated from 1968.1,2 In 1971 chloramphenicol resistance was reported in Salmonella Typhi detected in various Brazilian states3 and Shigella resistant to multiple antimicrobials were reported from Rio de Janeiro.4 At that time, β-lactam resistance was only reported for ampicillin.
The rise of extended-spectrum β-lactamasesThird generation cephalosporins became available for clinical use in Brazil in early 1980s. In our medical practice, we have observed resistance to third-generation cephalosporins among Enterobacteriaceae since 1985, but the first report of this finding in Brazilian hospitals was published only in 1994, describing a 52% cefepime resistance rate among ceftazidime-resistant Enterobacteriaceae.5 This was the first published clue on the presence of extended spectrum β-lactamases (ESBLs) in Brazil. In 1997, the first confirmation of ESBL production in Enterobacteriaceae from Brazil came out. The authors documented the presence of ESBLs in 72 K. pneumoniae clinical isolates, from private and public tertiary hospitals located in Rio de Janeiro and São Paulo, by clavulanic acid inhibition. Of note, they also reported a low susceptibility rates for amikacin (41.4%) and gentamicin (29.6%) but all isolates were still susceptible to imipenem.6 A subsequent work, also published in 1997, including 982 consecutive isolates from 18 hospitals, from four different states and seven different cities, was the first publication that could be used to estimate the ESBL rate among Enterobacteriaceae. Assuming that resistance to third generation cephalosporins was only due to ESBL production, 16% and 5% of K. pneumoniae and E. coli, respectively, would be classified as ESBL producers at that time.7
The first molecular studies on ESBLs from Brazil came out in 2000, evidencing the predominance of blaCTX-M genes and describing the CTX-M-8 enzyme in strains other than K. pneumoniae from Rio de Janeiro.8 The same group of researchers described the BES-1 and the CTX-M-16 enzymes in strains from the same city.9,10 Subsequent surveillance studies evidenced a growing ESBL production rates among Enterobacteriaceae collected from inpatients. In 2000, the ESBL production rate in K. pneumoniae collected from intensive care units was 59.2%, while these rates in Enterobacter spp. and E. coli were 19.5% and 14.6%, respectively.11 The most recent study on the diversity of ESBL types in Enterobacteriaceae isolated from Brazil refers to 1827 isolates collected during the period between August 2003 and March 2008 in the city of Curitiba, Paraná. CTX-M-2 was the most frequently detected ESBL in all Enterobacteriaceae species, except in Enterobacter aerogenes, in which a CTX-M-59-producing clone was predominant.12 Recent studies have reported that ESBL-producing Enterobacteriaceae are now detected in a significant rate in outpatients presenting cystitis. In a public institution located in Brasilia, the ESBL production rate in E. coli collected from July 2013 to April 2014 was 7.1%.13 When we reviewed all publication from Brazil about ESBLs, CTX-M-2 was the most frequently detected enzyme and was also detected in the largest number of different Enterobacteriaceae species (Table 1).
Extended spectrum β-lactamases detected in Enterobacteriaceae in Brazil.
| Enzyme | Species |
|---|---|
| BES-1 | Serratia marcescens9 |
| CTX-M-1 | K. pneumoniae14 |
| CTX-M-2 | Enterobacter aerogenes12,15; Enterobacter cloacae12,16; Escherichia coli12,16–22; Klebsiella pneumoniae14,16,21,23–29; K. oxytoca21; Morganella morganii16,21; Proteus mirabilis8,17,21; Providencia stuartii16,21,30; Salmonella typhimurium31; S. marcescens17 |
| CTX-M-3 | E. coli22 |
| CTX-M-8 | Citrobacter amalonaticus8; Enterobacter cloacae8; E. aerogenes8; E. coli12,16,22; K. pneumoniae14 |
| CTX-M-9 | Citrobacter freundii16; E. cloacae10,12; E. coli10,12,16,20; K. pneumoniae16 |
| CTX-M-14 | E. coli18,32 |
| CTX-M-15 | E. aerogenes12; E. cloacae12,33; E. coli18,22,32,34; K. pneumoniae27,33,35 |
| CTX-M-16 | E. coli10; E. cloacae10 |
| CTX-M-28 | K. pneumoniae24; K. oxytoca21 |
| CTX-M-59 | E. aerogenes15; E. cloacae15; E. coli20,21,27; K. pneumoniae21,23,27 |
| CTX-M-74 | E. cloacae16 |
| CTX-M-75 | P. stuartii16 |
| CTX-M-131 | P. stuartii36 |
| GES-1 | K. pneumoniae37 |
| GES-7 | K. pneumoniae38 |
| PER-2 | E. cloacae12,15 |
| SHV-2 | K. pneumoniae; E. coli22 |
| SHV-4 | K. pneumoniae39 |
| SHV-5 | E. cloacae16; E. coli16,19,21; K. pneumoniae16 |
| SHV-12 | E. aerogenes12,15; E. cloacae12,15; K. pneumoniae21,25,35 |
| SHV-27 | K. pneumoniae21,40 |
| SHV-28 | K. pneumoniae21,41 |
| SHV-31 | K. pneumoniae35 |
| SHV-38 | K. pneumoniae35 |
| SHV-40 | K. pneumoniae38 |
| SHV-45 | K. pneumoniae21 |
| SHV-55 | K. pneumoniae21 |
| SHV-108 | K. pneumoniae41 |
| SHV-122 | K. pneumoniae41 |
| TEM-15 | K. pneumoniae21 |
| TEM-115 | K. pneumoniae21 |
| TEM-116 | K. pneumoniae38 |
| TEM-135 | E. cloacae12 |
FOX-5-like and CMY-2-like were the first plasmid-mediated AmpCs (pAmpC) reported in Brazilian isolates. Both enzymes were detected in E. coli.42,43 The FOX-5-like encoding gene was detected during the DNA sequencing of a 41-kb conjugative plasmid that harbored a qnrA gene and a class 1 integron with the aadB and catB3 gene cassettes.43 The CMY-2-like enzyme was detected in four carbapenem-resistant E. coli strains, which also possessed alteration in the outer membrane proteins, isolated from a single patient.42 Dias and colleagues studied the prevalence of pAmpC among Enterobacteriaceae isolated from a teaching hospital in Rio de Janeiro. In that study pAmpC encoding genes were not detected and the multidrug resistance phenotype observed in five E. coli strain was attributed to hyperexpression of chromosomally encoded AmpC.44 Very few studies described the frequency of pAmpCs in Enterobacteriaceae in Brazil, although pAmpCs are of epidemiological importance, since carbapenem resistance can occur in strains with concomitant permeability alterations and pAmpC expression. A study conducted in a public tertiary hospital from São Paulo included 41 E. coli, five Klebsiella oxytoca, 65 Klebsiella pneumoniae, 18 P. mirabilis, and four Salmonella spp. detected during the period from January and July 2006 and found a 0.75% plasmid-mediated AmpC production rate and a single isolate producing a CMY-2-like enzyme was identified.45 The most recent study on pAmpCs in Brazil evaluated the frequency of plasmid-mediated AmpCs in E. coli isolated from urine cultures from both outpatients and inpatients. The frequency of plasmid-mediated AmpC was 0.46% in outpatients and 1.8% in inpatients. The full nucleotide sequences were determined and blaCMY-2 was the most frequently detected gene, but blaCMY-4 was also detected.46
The rise of carbapenemases in BrazilImipenem has been available for clinical use in Brazil since the end of the 80s. In 1989 a surveillance study, carried out with 1231 isolates, mainly from inpatients from five different medical centers from São Paulo, Rio de Janeiro and Salvador, reported a 1% imipenem resistance rate for E. coli, while this rate was 6% for Enterobacter sp. but resistant isolates were also found among K. pneumoniae.47 In 1998, a cefpirome susceptibility study also reported imipenem resistance in Enterobacteriaceae. Using commercial microdilution plates, 349 Enterobacteriaceae from tertiary hospitals from four different states were evaluated.48 Imipenem resistance rate among Enterobacter species varied from 2 to 8%, while this rate among K. pneumoniae was 7%, but unfortunately, no further studies were published on those strains. In 2005, almost 20 years after the introduction of carbapenems in clinical use in Brazil, the first report of an Enterobacteriaceae producing a carbapenemase came out. The work of Lincopan et al. described the presence of IMP-1 in a K. pneumoniae strain detected in a patient from a university hospital from São Paulo, located at the Southeast of Brazil.49 The same IMP-1-producing K. pneumoniae strain was detected in six distinct hospitals of the city of São Paulo between the years 2003 and 2005.50 In one of these hospitals, it was responsible for causing an outbreak in an intensive care unit.51 IMP-1 was also detected in a P. rettgeri isolate that was co-producer of CTX-M-like, and SHV-like.50 The co-production of IMP-10 and KPC-2 was detected in S. marcescens causing an outbreak in a tertiary, teaching hospital in Dourados, MS.52
GES-5, an enzyme of the GES family with spectrum toward carbapenems, was initially detected in Brazil in a K. pneumoniae isolated from a rectal surveillance swab of an elderly patient admitted to a private hospital in São Paulo, in 2008. This isolate also showed deletions on ompK35 and ompK36 genes.53 GES-5 was also detected in three genetically related Kluyvera intermedia that were isolated from one sink and two distinct taps of an intensive care unit of a tertiary-care hospital in Porto Alegre, in May 2013, during an environmental surveillance for NDM-1-producing isolates.54 Although only reported in 2014, GES-5 was also recovered from the blood of an adult patient admitted to a university hospital in Porto Alegre, in 2011. The patient had acute myeloid leukemia and had recent exposure to multiple antibiotics.55
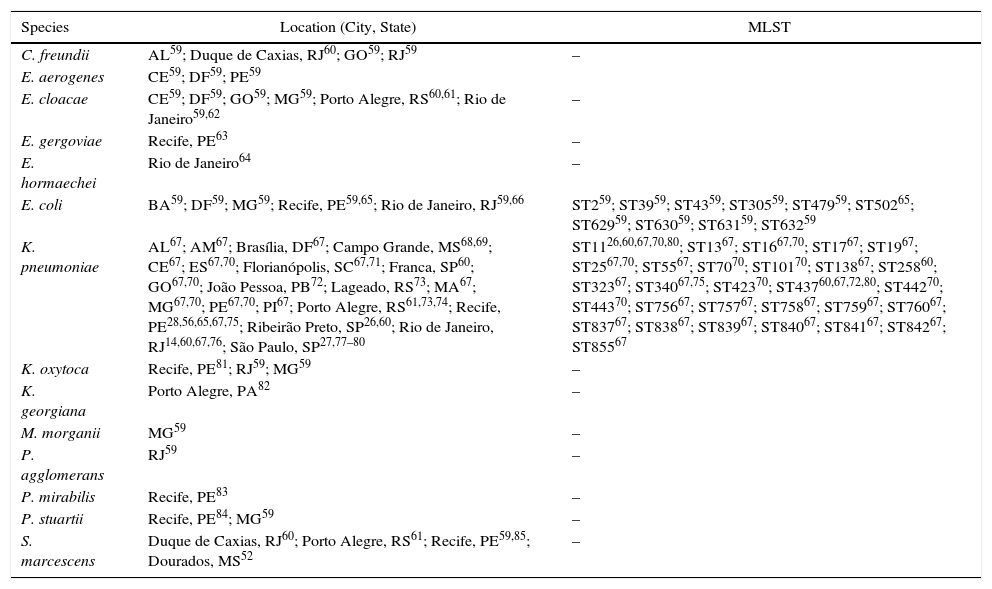
The first report on the detection of Klebsiella pneumoniae carbapenemase (KPC) in Brazil was published in 2009,56 and described the detection of KPC-2 in 2006, ten years after the first detection of KPC-2 in world, in 1996, in North Carolina, in the United States of America.57,58 The work described the detection of KPC-2 in K. pneumoniae in four patients from the city of Recife, located at the Northeast of Brazil. Earlier dissemination of KPC-2 production was later reported in São Paulo. Subsequently, KPC-2 was described in many Enterobacteriaceae species and locations all over Brazil (Table 2). To date this is the only variant reported from Brazil, although 23 variants (http://www.ncbi.nlm.nih.gov/pathogens/beta-lactamase-data-resources/) have been described worldwide.
KPC-producing species detected in Brazil.
| Species | Location (City, State) | MLST |
|---|---|---|
| C. freundii | AL59; Duque de Caxias, RJ60; GO59; RJ59 | – |
| E. aerogenes | CE59; DF59; PE59 | |
| E. cloacae | CE59; DF59; GO59; MG59; Porto Alegre, RS60,61; Rio de Janeiro59,62 | – |
| E. gergoviae | Recife, PE63 | – |
| E. hormaechei | Rio de Janeiro64 | – |
| E. coli | BA59; DF59; MG59; Recife, PE59,65; Rio de Janeiro, RJ59,66 | ST259; ST3959; ST4359; ST30559; ST47959; ST50265; ST62959; ST63059; ST63159; ST63259 |
| K. pneumoniae | AL67; AM67; Brasília, DF67; Campo Grande, MS68,69; CE67; ES67,70; Florianópolis, SC67,71; Franca, SP60; GO67,70; João Pessoa, PB72; Lageado, RS73; MA67; MG67,70; PE67,70; PI67; Porto Alegre, RS61,73,74; Recife, PE28,56,65,67,75; Ribeirão Preto, SP26,60; Rio de Janeiro, RJ14,60,67,76; São Paulo, SP27,77–80 | ST1126,60,67,70,80; ST1367; ST1667,70; ST1767; ST1967; ST2567,70; ST5567; ST7070; ST10170; ST13867; ST25860; ST32367; ST34067,75; ST42370; ST43760,67,72,80; ST44270; ST44370; ST75667; ST75767; ST75867; ST75967; ST76067; ST83767; ST83867; ST83967; ST84067; ST84167; ST84267; ST85567 |
| K. oxytoca | Recife, PE81; RJ59; MG59 | – |
| K. georgiana | Porto Alegre, PA82 | – |
| M. morganii | MG59 | – |
| P. agglomerans | RJ59 | – |
| P. mirabilis | Recife, PE83 | – |
| P. stuartii | Recife, PE84; MG59 | – |
| S. marcescens | Duque de Caxias, RJ60; Porto Alegre, RS61; Recife, PE59,85; Dourados, MS52 | – |
The following abbreviations correspond to Brazilan States: AL, Alagoas; AM, Amazonas; BA, Bahia; DF, Distrito Federal; ES, Espírito Santo; GO, Goiás; MA, Maranhão; MG, Minas Gerais; MS, Mato Grosso do Sul; PB, Paraíba; PE, Pernambuco; RJ, Rio de Janeiro; RS, Rio Grande do Sul; SC, Santa Catarina; SP, São Paulo.
KPC-2-producing Enterobacteriaceae are now disseminated all over Brazil but K. pneumoniae is the most frequent species. Among this species, ST11 and ST437, which belong to the clonal complex 258, are the most frequently detected clonal groups (Table 2). A recent publication analyzed 3085 K. pneumoniae isolates cultivated from patients from 10 private hospitals from the great São Paulo urban area, during the period from January 2011 to December 2015. Most of the isolates were recovered from blood cultures. The work showed an amazing increase in the carbapenem resistance rate, from 6.8% in 2011 to 35.5% in 2015; of note, KPC-2 was detected in 96.2% of the carbapenem-resistant isolates, and there were both interhospital and intrahospital clonal dissemination.80
New Delhi metallo-β-lactamase (NDM) was firstly detected in Brazil in 2013, in Providencia rettgeri, from a patient from Porto Alegre, a city located at the South of Brazil.86 This detection occurred four years after the initial detection in a ST14 K. pneumoniae strain causing urinary tract infection and in an E. coli strain from feces from a Swedish patient of Indian origin.87 Compared to what happened with KPC, which was detected in Brazil 10 years after the initial description, NDM-1-producing strains were detected much earlier, which indicates a great potential for more efficient dissemination in Brazil. This P. rettgeri strain was later shown to have heterogeneous carbapenem resistance, which could make its detection a challenging task.88 Subsequently, expression of NDM-1 was reported in E. hormaechei from the same city where the first case had been detected.89E. cloacae complex strains and Morganella morganii expressing NDM-1 were also reported by other research group from Porto Alegre.90 In the same year of 2013, the first complete nucleotide sequences of blaNDM-1-bearing plasmids from Brazil were described, in E. coli and E. hormaechei cultured from the same rectal swab from a patient from Rio de Janeiro who had never been exposed to carbapenems. The blaNDM-1 gene was found to be located on a IncFIIk in E. hormaechei and on a IncX3 plasmid in a ST2 E. coli, but both plasmids contained a new structure designated Tn3000, that could possibly mediate the transposition of the blaNDM-1 gene.91 Subsequently, coproduction of NDM-1 and KPC-2 was described in P. rettgeri and E. cloacae from Rio de Janeiro.62,64
More recently, a new class A carbapenemase, designated Brazilian Klebsiella carbapenemase (BKC-1) was described in Brazil.92 To date it has only been found in K. pneumoniae in a low frequency, possible due to the fact that the blaBKC-1 gene is located in a small transferable, non-conjugative plasmid.92,93
Polymyxin resistance in carbapenem-resistant K. pneumoniae: a nightmareThe first report on polymyxin-resistance in Brazilian Enterobacteriaceae came out in 2006.94 At that time, with a low colistin and polymyxin B clinical use, the polymyxin B resistance rate was 0.5% in E. coli, 1.8% in K. pneumoniae and 16.7% in Enterobacter spp. In a subsequent publication the same group evidenced a 3.0% resistance rate among K. pneumoniae in Latin America.95 In 2013, Pereira et al. reported a 15% polymyxin resistance rate among KPC-producing K. pneumoniae from diverse Brazilian states.67 Polymyxin resistance in E. cloacae and K. pneumoniae strains was also reported from Porto Alegre, south of Brazil.61,96 To date, interruption of mgrB gene by insertion sequences or missense mutations is the most frequent mechanism of polymyxin resistance in K. pneumoniae in Brazil.93,97 The most recent evidence of the amazing polymyxin resistance problem in Brazil came from a report from São Paulo, the largest city in Latin America. The authors described a 35.5% carbapenem resistance rate due to KPC-2 production, among K. pneumoniae causing infections in 2015, and also found an increase in polymyxin B resistance among KPC-producing K. pneumoniae, from 0% in 2011 to 27.1% in 2015.80 This increase coincided with the increased use of polymyxins as empiric therapy to treat severe infectious when Gram negatives are possible etiologic agents in intensive care units. More concerning was finding interhospital and intrahospital clonal dissemination and the fact that most isolates included in the study were detected from blood cultures. This is a therapeutic nightmare, since intravenous fosfomycin and ceftazidime-avibactam are still not available in Brazil.
Recently, the mcr-1 (mobile colistin resistance) gene was detected in a clinical strain of E. coli ST101 from the Northeast of Brazil.98 The authors found the gene located on a IncX4 plasmid, and consequently the selective pressure represented by the overuse of polymyxins could have contributed to the dissemination of this resistance mechanism.
In summary, antimicrobial resistance in Enterobacteriaceae in Brazil is a very serious problem that needs urgent actions which includes more strict adherence to infection control measures, more judicious use of antimicrobials in human and animal husbandries and fast approval of old and new antimicrobials like fosfomycin and ceftazidime-avibactam for clinical use in Brazil, in order to decrease the polymyxin consume. If these measures are not applied together, the release of ceftazidime-avibactam will be a partial measure that will be probably followed by dissemination of NDM-1 producers in Brazil.
Conflicts of interestThe authors declare no conflicts of interest.