Nowadays, it is necessary to search for different high-scale production strategies to produce recombinant proteins of economic interest. Only a few microorganisms are industrially relevant for recombinant protein production: methylotrophic yeasts are known to use methanol efficiently as the sole carbon and energy source. Pichia pastoris is a methylotrophic yeast characterized as being an economical, fast and effective system for heterologous protein expression. Many factors can affect both the product and the production, including the promoter, carbon source, pH, production volume, temperature, and many others; but to control all of them most of the time is difficult and this depends on the initial selection of each variable. Therefore, this review focuses on the selection of the best promoter in the recombination process, considering different inductors, and the temperature as a culture medium variable in methylotrophic Pichia pastoris yeast. The goal is to understand the effects associated with different factors that influence its cell metabolism and to reach the construction of an expression system that fulfills the requirements of the yeast, presenting an optimal growth and development in batch, fed-batch or continuous cultures, and at the same time improve its yield in heterologous protein production.
The need for new ways to produce recombinant proteins of economic interest has been oriented toward culture processes in bioreactors under controlled conditions to reduce production costs and simplify and facilitate its acquisition.1
The methylotrophic yeast Pichia pastoris (Komagataella phaffii) is one of the most commonly used expression systems for heterologous protein production.2 Recombinant protein obtained in a P. pastoris system exhibits one improved aspect: the specific activity of the enzyme produced has been shown to have increased when this was assessed. This is highly relevant because when a recombinant protein is destined for industry use, product yield is an important factor in the profitability of the process.3 However, the growth of yeast in culture medium is affected by such factors as: (a) methanol concentration, which one is used to induce the heterologous protein production, but the toxic and inflammatory nature of methanol restricts its application, especially in edible and medical products4; (b) carbon sources that can influence both cell and recombinant protein production either in flasks or in a bioreactor5; (c) oxygen concentration, which can generate aerobic or anaerobic environments; (d) temperature, which affects important cell processes, such as central carbon metabolism, stress response and protein folding6; (e) high protease expression levels; (f) nutrient-deficiency when grown in defined media; (g) difficulties in systematic study due to product-specific effects; and (h) health and safety concerns associated with the storage of large quantities of methanol. Difficulties also arise that are specific to the culture methods and control strategies used.7 All these variations in metabolism produced by cellular and molecular responses to the media that are highly variable in terms of the growth stability of the recombinant yeast limit the recombinant protein production process at some point, resulting in a process with a lower yield and higher costs. Protein stability is a topic of major interest for the biotechnology, pharmaceutical and food industries, and it may primarily consider the half-life of a protein's activity. An understanding of protein stability is essential for optimizing the expression, purification, formulation, storage and structural studies of proteins.66,67 There is evidence that demonstrate that the residues flanked by well-conserved segments among homologous sequences tended to contribute to the stability of the protein to a greater extent than did residues flanked by less conserved segments) glycosytation stage: better energy interactions have been observed in the glycosylated protein compared to the non-glycosylated, affecting substrate affinity and stability that.68,69 Therefore, it is necessary to know the main factors to control the culture conditions throughout the process to produce high concentrations of recombinant proteins. This review focuses on two points: the selection of the best promoter in the recombination process considering different carbon sources as inductors, and the temperature as a culture medium variable that has been shown to be related to the uptake of the substrate (inductor) on methylotrophic Pichia pastoris yeast to understand the effects on its cell metabolism and to reach the construction of an expression system that fulfills the requirements of the yeast, presenting an optimal growth and development in batch, fed-batch or continuous cultures, and at the same time improve its yield in heterologous protein production.
Methylotrophic yeastEukaryotic methylotrophs, which are able to obtain all the carbon and energy needed for growth from methanol, are restricted to a limited number of yeast species. When these yeasts are grown on methanol as the sole carbon and energy source, the enzymes involved in methanol metabolism are strongly induced, and the membrane-bound organelles, peroxisomes, which contain key enzymes of methanol metabolism, proliferate massively. When cells grown in methanol are transferred to culture media containing different carbon sources, such as glucose or ethanol, the peroxisomes quickly disappear as a result of active degradation, which involves the proteolytic degradation of the peroxisomes.8 Like other yeasts, methylotrophic yeasts can use glycerol, ethanol and acetate.5,9,10
Complete oxidation of methanol to CO2 and H2O in methylotrophic microorganisms includes both dissimilatory pathways (energy generation) and assimilatory pathways (biosynthesis of cell material). However, this metabolism in methylotrophic yeasts differs from that of methylotrophic bacteria in three main respects11:
- (a)
The nature of the enzymes involved: Yeasts do not have methanol dehydrogenases for methanol oxidation, but rather alcohol oxidases.
- (b)
Compartmentalization of the process: In yeasts, part of the methanol oxidation occurs within the cell organelles called peroxisomes. These membranous compartments have some of the enzymes and metabolites involved in the methanol oxidation process, such as alcohol oxidase and catalase. During growth in the presence of methanol, peroxisomes can occupy 90% of the cell volume, whereas in the presence of other carbon sources, like glucose or glycerol, they are undetectable.
- (c)
The energy generation pathway: In methylotrophic bacteria, methanol oxidation is linked to the electron transport chain. However, in methylotrophic yeasts, reducing power is produced in the form of NADH in reactions once the methanol has been fixed in formaldehyde. This NADH is produced in the cytosol, oxidized in the absence of specific transporters, and moves toward the interior of the mitochondrion due to the activity of a NADH dehydrogenase located on the outside of the internal mitochondrial membrane.
The main application for methylotrophic yeasts, or at least the most important in the biotechnological sphere, was the discovery of their exceptional ability to act as host cells to heterologous proteins.8 The genera Pichia and Hansenula are interesting and advantageous alternatives to conventional foreign protein expression systems, like Escherichia coli or the commonly known bread yeast Sacharomyces cerevisiae.12
Pichia pastoris yeastPichia pastoris is one of the most important hosts for recombinant protein production in the biotechnological industry, mainly related to pharmaceutical production.13 This yeast was first described by Guilliermond in 1919. Its morphology is highly variable, able to present spherical or oval-shaped cells, single or combined in pairs. However, the cells can alter their shape according to the culture conditions; for example, they are frequently oval in favorable growth conditions but under no circumstances form pseudohyphae or hyphae. In solid medium they form white or cream-colored non-filamentous colonies. Under a microscope several buds can be observed.14
P. pastoris is anaerobic, facultative and methylotrophic, which means that it metabolizes methanol as a carbon and energy source. This capacity is conferred by the AOX (alcohol oxidase) gene, which is responsible for the induction of the enzyme alcohol oxidase, the enzyme that catalyzes the oxidation of methanol to formaldehyde and hydrogen peroxide. In the yeast genome, the location of the AOX gene is used to insert a vector. This vector contains a heterologous gene, a selectable marker gene (histidine dehydrogenase gene) and a secretion signal (α factor).15–18 Thus, adding methanol leads to the expression of the heterologous gene responsible for the synthesis of recombinant proteins, such as antibody fragments.4
For heterologous protein production in cultures in a fermenter, a high-density three-step fermentation scheme is normally used. In the first stage, the recombinant yeast is cultured in a saline medium with a non-fermentable carbon source, such as glycerol. Once the glycerol is depleted, the second phase (transition phase) is begun by adding glycerol at a limiting growth rate. The second phase is important because the by-products generated (for example, ethanol) during the batch phase are consumed and the cells are prepared for induction. The third phase (induction phase) is started by adding limited concentrations of methanol, which results in recombinant protein production.16 After being transferred to the medium with methanol, this organism undergoes a massive proliferation of peroxisomes while they induce the synthesis of the more than 10 enzymes needed for methanol metabolism, including the peroxisomal enzyme AOX.8 The strong induction of protein synthesis by methanol in P. pastoris has been used as an ideal system of different heterologous gene expression, as can be seen in Table 1; moreover, this yeast has become one of the most important microorganisms, allowing heterologous protein expression by secretion within the supernatant of the (extracellular) culture or by intracellular localization.13,19 This model of expression makes it possible to obtain from milligrams to grams of functionally active recombinant protein, which is used in basic research and therapeutic administration (Table 1). This system has several characteristics in addition to high growth densities and productivity. These characteristics are:
- (a)
The cost of the media, equipment and infrastructure of the yeast cultures are much more economical than those from cells from mammals and insects.13
- (b)
The protein expression system designed could be induced by adding methanol, as an alternative for the production of recombinant proteins in P. pastoris, and it is generally integrated into the chromosome of the yeast to gain stability in the genetic construct.20
- (c)
This microorganism makes post-translational modifications like glycosylation, adding a small sugar pattern very similar to the eukaryotic cells. For this reason, methylotrophic yeasts such Pichia pastoris and Hansenula polymorpha are preferred as the overall length of the mannose outer chains is shorter than in S. cerevisiae.21,22
- (d)
The possibility of directly secreting the cloned protein, which in practical terms means a pre-purification step.13,22
- (e)
This yeast does not produce endotoxins; therefore, it may be considered a safe microorganim.22
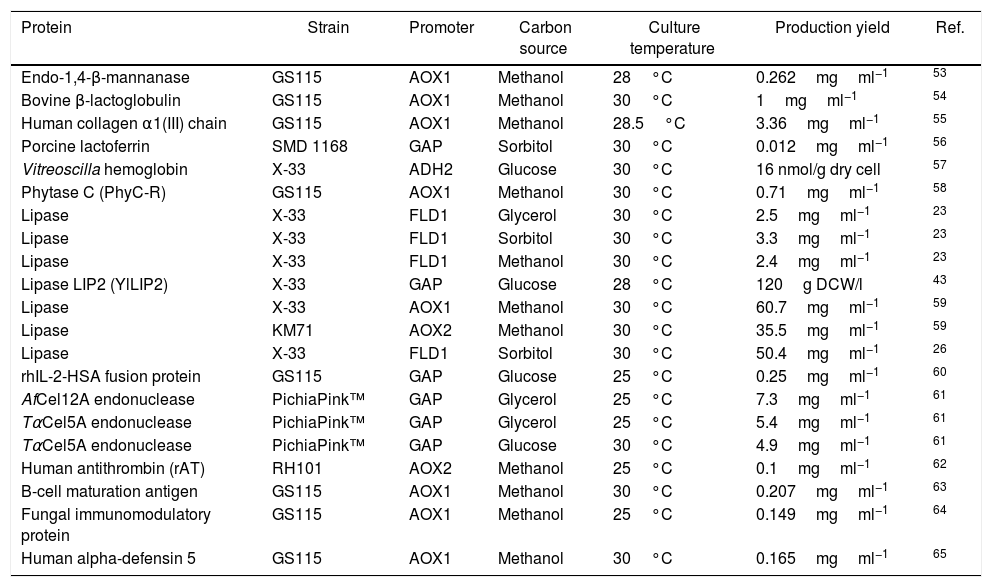
Production of heterologous proteins in different strains of the yeast Pichia pastoris, indicating the temperature and carbon source used in the induction phase, the promoter and the protein yield produced.
| Protein | Strain | Promoter | Carbon source | Culture temperature | Production yield | Ref. |
|---|---|---|---|---|---|---|
| Endo-1,4-β-mannanase | GS115 | AOX1 | Methanol | 28°C | 0.262mgml−1 | 53 |
| Bovine β-lactoglobulin | GS115 | AOX1 | Methanol | 30°C | 1mgml−1 | 54 |
| Human collagen α1(III) chain | GS115 | AOX1 | Methanol | 28.5°C | 3.36mgml−1 | 55 |
| Porcine lactoferrin | SMD 1168 | GAP | Sorbitol | 30°C | 0.012mgml−1 | 56 |
| Vitreoscilla hemoglobin | X-33 | ADH2 | Glucose | 30°C | 16 nmol/g dry cell | 57 |
| Phytase C (PhyC-R) | GS115 | AOX1 | Methanol | 30°C | 0.71mgml−1 | 58 |
| Lipase | X-33 | FLD1 | Glycerol | 30°C | 2.5mgml−1 | 23 |
| Lipase | X-33 | FLD1 | Sorbitol | 30°C | 3.3mgml−1 | 23 |
| Lipase | X-33 | FLD1 | Methanol | 30°C | 2.4mgml−1 | 23 |
| Lipase LIP2 (YlLIP2) | X-33 | GAP | Glucose | 28°C | 120g DCW/l | 43 |
| Lipase | X-33 | AOX1 | Methanol | 30°C | 60.7mgml−1 | 59 |
| Lipase | KM71 | AOX2 | Methanol | 30°C | 35.5mgml−1 | 59 |
| Lipase | X-33 | FLD1 | Sorbitol | 30°C | 50.4mgml−1 | 26 |
| rhIL-2-HSA fusion protein | GS115 | GAP | Glucose | 25°C | 0.25mgml−1 | 60 |
| AfCel12A endonuclease | PichiaPink™ | GAP | Glycerol | 25°C | 7.3mgml−1 | 61 |
| TαCel5A endonuclease | PichiaPink™ | GAP | Glycerol | 25°C | 5.4mgml−1 | 61 |
| TαCel5A endonuclease | PichiaPink™ | GAP | Glucose | 30°C | 4.9mgml−1 | 61 |
| Human antithrombin (rAT) | RH101 | AOX2 | Methanol | 25°C | 0.1mgml−1 | 62 |
| B-cell maturation antigen | GS115 | AOX1 | Methanol | 30°C | 0.207mgml−1 | 63 |
| Fungal immunomodulatory protein | GS115 | AOX1 | Methanol | 25°C | 0.149mgml−1 | 64 |
| Human alpha-defensin 5 | GS115 | AOX1 | Methanol | 30°C | 0.165mgml−1 | 65 |
The heterologous protein expression system in P. pastoris is based mainly on the use of the alcohol oxidase enzyme PAOX1 promoter, which is heavily regulated and induced by methanol23; however, there are several parameters that can affect this expression system. Some are intrinsic to the expression system, for example the Mut phenotype of the strain, the insertion site of the gene of interest in the cell genome, the number of copies of the foreign gene, the type of intra or extracellular expression, the nature of the secretion signal (in case of extracellular expression) or the activity of endogenous proteases. Other factors are related to the protein in question to be expressed, like the GC/AT content of the gene to be expressed or the degree of toxicity of the foreign protein for the host cell. Also, other important factors are the stability of its mRNA, as well as the post-transcriptional and/or translational modifications required.12
Influence of the carbon source on the cultureMethanol metabolism in P. pastorisThe cytosolic and peroxisomal enzymes involved in methanol metabolism are synthesized when P. pastoris grows in the presence of methanol. When cells adapted to grow in methanol are transferred to a medium with glucose or ethanol, these enzymes are sequestered selectively and quickly, and degraded in the vacuoles of the yeast.10
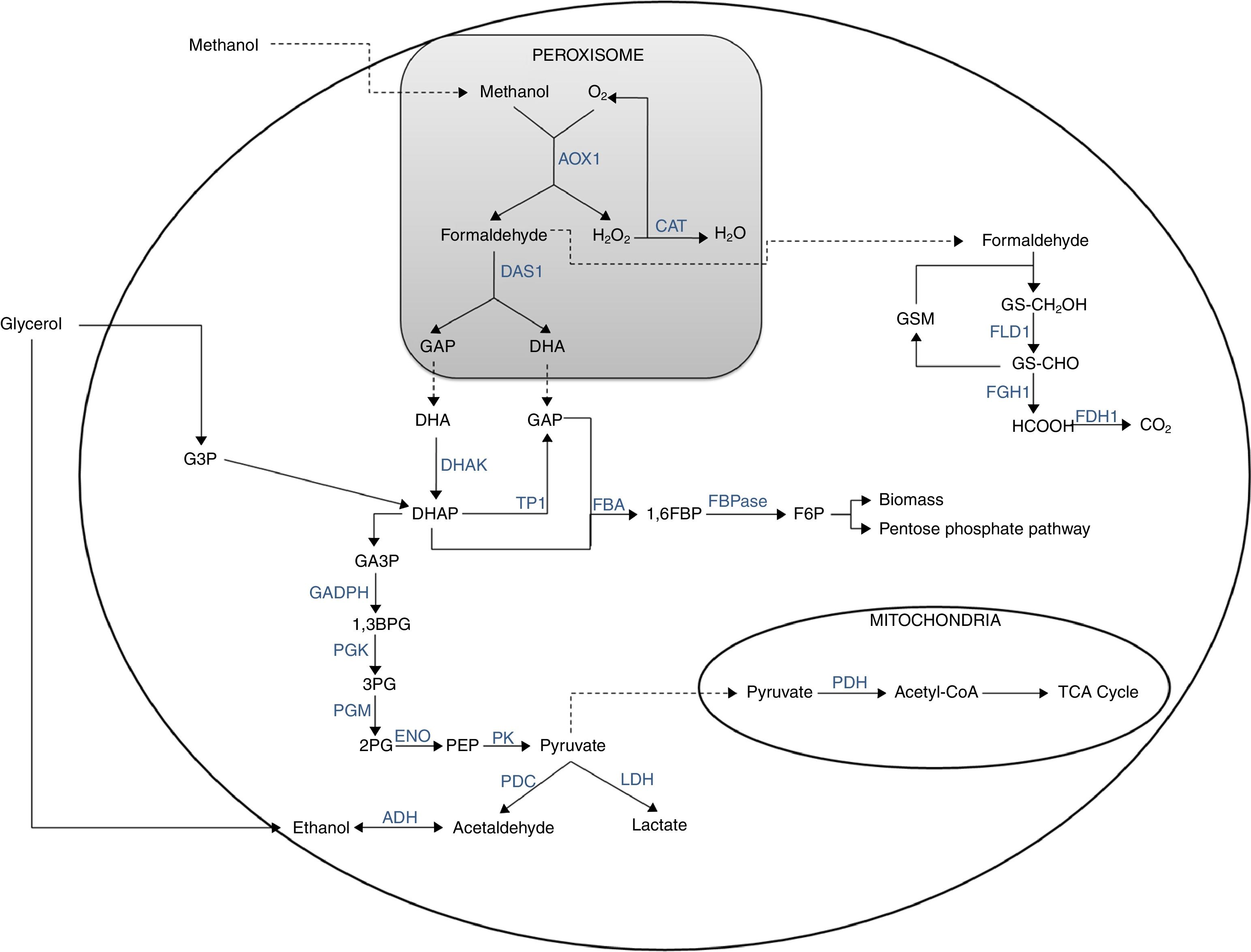
Methanol metabolism begins inside the peroxisomes (Fig. 1). The metabolism begins when the methanol is oxidized in the presence of oxygen by the enzyme alcohol oxidase, producing formaldehyde and H2O2. Hydrogen peroxide is toxic to the cell; therefore, it is retained in the peroxisome until the catalase converts it into H2O and O2. Formaldehyde through the action of the transketolase dihydroxyacetone synthase (E.C.2.2.1.3) enzyme is turned into dihydroxyacetone (DHA) and glyceraldehyde-3-phosphate (GA3P). DHA is released to the cytosol and phosphorylated by dihydroxyacetone kinase (E.C. 2.7.1.29) enzyme to form dihydroxyacetone phosphate (DHAP). DHAP can undergo two reactions: (1) triose phosphate interconversion through the action of the triose phosphate isomerase (E.C. 5.3.1.1) enzyme to form GA3P and, therefore, to feed the second phase on the glycolytic pathway; and (2) the reaction most frequently reported in the literature is the condensation of DHAP with a glyceraldehyde phosphate (GAP) molecule to form fructose 1,6 bisphosphate (1,6FBP) and then fructose 6-phosphate (F6P). In these transformations the enzymes fructose 1,6 bisphosphate aldolase (FBA, E.C.4.1.2.13) and fructose 1,6 bisphosphate (FBPase, E.C.3.1.3.11) intervene respectively; the latter is part of the process known as gluconeogenesis. Alternatively, F6P can form biomass or be incorporated into the pentose phosphate pathway (pentose pathway) to form xylulose 5-phosphate (Xu5P) and resupply the cycle in DHAP; likewise, DHAP is regenerated several times, which favors biomass formation. The remaining formaldehyde from the methanol oxidation flows into the cytosol, where it reacts with glutathione through the enzymes formaldehyde dehydrogenase (E.C.1.2.1.1) and formate dehydrogenase (E.C.1.2.1.2) to form CO2 with detachment of NADH+H (energy source).24,25
It is noteworthy that the initial oxidation of methanol to formaldehyde does not generate any type of metabolic energy (either in the form of reducing power, NADH, or in the form of ATP), strongly affecting (negatively) the growth performance of these microorganisms with respect to methanol and oxygen.24
There is evidence that P. pastoris can grow in three carbon sources (glycerol, methanol and ethanol) as a sole carbon and energy source. Diauxic growth has been observed when two of the three carbon sources are present, particularly in ethanol-glycerol and ethanol-methanol mixtures, with glycerol being preferred over ethanol and methanol, and ethanol being preferred over methanol. In the first case, once all the glycerol has been consumed, the use of ethanol is accompanied by a temporary accumulation of acetate, which later served as a carbon source, whereas in the second case, this accumulation is very low. Another important aspect is that in cultures in presence of glycerol and methanol a diauxic growth is not observed due to methanol begins to be consumed before glycerol has been spent from the medium. It is worth noting that most of the enzymes involved in methanol metabolism differ to those of glycerol. Otherwise, when more of one carbon source is present, the order of use is: glycerol, ethanol, acetate (which is accumulated due to the use of ethanol) and methanol.16
Ethanol metabolism in P. pastorisAlcoholic fermentation, which depends on the respiratory capacity and metabolic rate of the glucolysis, can be triggered from pyruvate (Fig. 1). In this process, pyruvate decarboxylase (E.C.4.1.1.1) transforms pyruvate into acetaldehyde and the latter is reduced to form ethanol, a reaction catalyzed by the enzyme alcohol dehydrogenase (E.C.1.1.1.1). In addition, ethanol can be used as a carbon and energy source if the respiratory capacity of the yeast is re-established. In this case, ethanol would be transformed into acetaldehyde by the alcohol dehydrogenase and then into acetate by the acetaldehyde dehydrogenase (E.C. 1.2.1.10) and finally the acetyl-CoA synthase converts it to acetyl-CoA.24,25
Glycerol is a non-fermentable carbon source and P. pastoris is not considered a fermentative yeast; however, ethanol is accumulated during the culture when is used glycerol at a high feed rate. This phenomenon has been reported on several occasions but the mechanism has yet to be explained. Some authors have considered that ethanol is metabolized to acetaldehyde and then to acetate, which is assimilated as acetyl-CoA, a process which has been described in H. polymorpha and for P. methanolica. The regulation of the methanol metabolism in methylotrophic organisms is a complex process that includes the synthesis, activation and degradation of the enzymes involved. These enzymes are induced by methanol, formaldehyde and formate (positive effectors). However, the activity of the enzyme alcohol oxidase is reduced by glucose and ethanol (negative effectors) through two regulation mechanisms: catabolite repression and inactivation. The first of these mechanisms involves the control over RNA synthesis, and the second involves enzyme inactivation or degradation. Acetate formation as a result of the use of ethanol represses the expression of the alcohol oxidase while it is depleted.16
Respiro-fermentative metabolismIt should be noted that P. pastoris is a microorganism that prefers a respiratory metabolism, thereby avoiding the typical undesirable by-product generation of fermentative processes, like ethanol, which facilitates its culture at high cell densities (>100g of weight perL−1). Reaching high biomass levels is highly desirable in the expression of proteins associated with cell growth.26
The high cell densities in P. pastoris yeast are related to a change from a respiratory metabolism to a respiro-fermentative one.27,28 This is very similar to what has been observed in the metabolism of cancer cells, where a high rate of proliferation is observed that generates genetic changes related to the increased metabolism. Another important reason for a change in metabolism is the adaptation of tumor cells to the microenvironment. Hypoxic areas are frequently found due to the rapid tumor growth. In conditions of severe hypoxia, the cells are forced into an anaerobic glycolysis as their primary energy source (the Pasteur effect).29 Effects of oxygen transfer on recombinant protein production by Pichia pastoris under GA3P promoter producing recombinant glucose isomerase were investigated by Güneş and Çalık.30 Two groups of oxygen transfer strategies were applied, one of which was based on constant oxygen transfer rate where the aeration rate (QO/V)=3 and 10vvm, and the agitation rate was N=900min−1; the other one was based on constant dissolved oxygen concentrations (CDO)=5, 10, 15, 20 and 40% in the fermentation broth, by using predetermined exponential glucose feeding with μo=0.15h−1. The highest cell concentration was obtained as 44gL−1 at t=9h of the glucose fed-batch phase at CDO=20% operation while the specific enzyme activities revealed that keeping CDO at 15% was more advantageous at the expense of relatively higher by-product formation and lower specific cell growth rate.
Some promoters used to induce heterologous proteinsAlcohol oxidaseThe enzyme AOX (EC 1.1.3.13) belongs to the family of glucose-methanol-choline oxidoreductases, catalyzing the oxidation of short aliphatic alcohols, such as methanol, ethanol and 1-propanol. It is mainly found in the peroxisomal matrix of methylotrophic yeasts. The active mature form of AOX is an oligomer: AOX in some methylotrophic yeast species is a molecule of high molecular weight (600kDa) that consists of eight identical subunits, each of which have as a prosthetic group a non-covalently bonded flavin adenine dinucleotide (FAD) molecule, although tetrameric and hexameric forms have also been found (31). This enzyme is encoded by the AOX1 gene, and presents low affinity for O2, with the methanol oxidation rate in cell suspensions being linearly proportional to the oxygen concentration dissolved in the medium. The catalytic activity of this enzyme is below its optimal conditions, which is why the organism compensates it with an increase in its synthesis rate in order to obtain high concentrations of the enzyme (Table 1).32
AOX is a key enzyme in methanol metabolism and catalyzes the first step in the catabolism of methanol: oxidation of methanol to formaldehyde with the resulting production of H2O2. It presents two isoenzymes, AOX1 and AOX2, which share 97% identity in the amino acid sequence, but their expressions are controlled by different promoters.31 There are three types of P. pastoris host strains available that differ from their ability to metabolize methanol and thus from their oxygen consumption rate. These are the Mut+ phenotype (wild-type strain, high oxygen consumption), MutS (methanol utilization slow, resulting from AOX1 deletion, intermediate oxygen consumption) and Mut− (methanol utilization minus, resulting from the double AOX1-AOX2 deletion, low oxygen consumption).11,33 The Mut+ phenotype is mainly used for recombinant protein productions, although the MutS phenotype has been used in some other cases. However, the use of methanol as the sole carbon source during the induction phase for protein production poses one major drawback for Mut+ strains (and MutS in a lower extent): as a high-degree reductant with a high heat of combustion, methanol catabolism requires a high oxygen consumption in aerobic conditions leading accordingly to a huge amount of heat production.34–36AOX1 constitutes the majority of the AOX protein in the cell, and is the main contributor for methanol metabolism; the growth of a line with defective AOX1 (MutS) is very slow in the medium with methanol, whereas a defect in AOX2 has little effect on growth.31,33 In MutS strains the force of the AOX1 promoter can be directed mainly toward recombinant protein production instead of using energy for AOX1 protein production. Nevertheless, most researchers use a wild type strain, although some researchers showed that MutS strains were advantageous for production.35,37
Formaldehyde dehydrogenaseThe strength of the formaldehyde dehydrogenase (FLD1) promoter is comparable to the AOX promoter, which makes it an attractive alternative for recombinant protein production.38 This promoter has the flexibility to induce high expression levels using methanol or methylamine.39 In addition, it has been shown that the use of sorbitol as a carbon source combined with methylamine as a nitrogen source is the base for developing methanol-free fed-batch fermentation processes for the production of heterologous proteins in P. pastoris based on the FLD promoter.26
Although the cells have a great capacity to regenerate the cofactors (NADH and NAD), the cellular demand and high recombinant enzyme activity in combination with their high overexpression within the cell can produce a limitation of the cofactor for the biotransformation process.40 In this situation, FLD has had a large impact on improving the theoretical NADH formation rate, as this is a limiting step in the methanol dissimilation pathway. By contrast, both the increase in formate dehydrogenase (FDH) activity and the increase in the Vmax of AOX (over 0.3U/mg) were at a very low theoretical NADH formation rate; additionally, the formaldehyde generated as a by-product of the AOX pathway strongly inhibits FLD, resulting in NADH formation problems.41 On the other hand, compared with the AOX promoter, the FLD promoter has presented significantly better results in terms of yield, productivity and specific productivity in systems expressing the same protein.26
Glyceraldehyde-3-phosphate dehydrogenase (GAP)The GAP promoter is used for the constituent expression of several heterologous proteins30,42–44; moreover, based on the expression system, P. pastoris has been shown to improve the yield in the production of proteins compared to the methanol-inducible promoter AOX1. Added to this, use of this promoter does not require methanol use,45 which implies that it is not necessary to change the carbon source during the growth and induction process, meaning the culture is easier to implement,39 showing great potential for the execution of continuous cultures.45 Nevertheless, as this promoter is constituently expressed, it is not a good choice for the production of proteins that are toxic to the yeast.39
The activity of the GAP promoter in cells growing with glycerol and methanol are approximately two thirds and one third of the level observed for glucose, respectively.39 It must be considered that glucose is the most frequently used substrate for the regeneration of nicotinamide cofactors for cell metabolism.46 These are important in the amino acid and protein syntheses which impact on the levels of transcripts and/or proteins related to ribosome biogenesis and translation.47
Alcohol dehydrogenasesThe presence of two alcohol dehydrogenases (mitochondrial ADH and ADH3) has been observed at the end of the glycerol batch phase, indicating the formation of ethanol, which was confirmed by gas chromatography (1.3gL−1 of ethanol). Previous research has also documented ethanol formation by P. pastoris during growth in excess glycerol. Surprisingly, the number of both enzymes increased even more in the methanol fed-batch phase (0.3gL−1), indicating possible participation in metabolic activities beyond the ethanol metabolism. In fact, it has been suggested that the proteins in the ADH family of other methylotrophic yeasts (e.g., C. boidinii) are involved in detoxification by formaldehyde by means of methyl formate formation. A similar role can also be attributed to ADHs in P. pastoris.48
Recombinant protein production under the control of the ADH3 promoter was compared to both AOX1 promoter and GAP promoter in Pichia pastoris expressing the Aspergillus niger xylanase (XylB) gene initiated by adding ethanol, methanol and glucose, respectively in the culture medium, where extracellular protein production yield for ADH3 promoter (3725U/mL) was higher than for AOX1 promoter (2095U/mL) and GAP promoter (580U/mL) at fermenter scale carried out for 72h at 30°C, pH 5 and 30% dissolved oxygen.9
Influence of temperature on culturesThere is evidence that the biggest bottleneck in terms of heterologous protein secretion is protein folding and cell secretion machinery due to the secretion of proteases into the medium, possibly due to increased cell lysis, causing a rise in proteolytic degradation, which is a significant problem in many high cell-density cultures.6,49 Previous studies have reported that the decrease in growth temperature can lead to an improvement in the production of batch, fed-batch and continuous culture systems.47,50 Concerning the positive effect of the reduction in growth temperature on specific heterologous protein productivity, it is believed that at least during the methanol cultures, a reduction in cell lysis and proteolytic activity is responsible for the high yield of culture production at low temperatures, although no detailed studies as yet exist in this regard.6
Use of the temperature-limited fed-batch (TLFB) technique is an alternative fed-batch technique in which the common methanol limitation is replaced by temperature limitation in order to avoid oxygen limitation at high cell density. This technique has resulted in higher cell density, lower cell death, higher concentration of the product and drastically lower proteolytic degradation of the recombinant protein compared to corresponding methanol limited P. pastoris bioreactor cultures. It has been observed that working on cultures with low temperatures (12°C), and considering that all the substrates present are not in limiting conditions, the dissolved oxygen tension (DOT) can be kept constant, while the cells are maintained at their maximum growth rate (μ) at all times, which enables the acquisition of a high biomass. The existing relation between growth rate and temperature is usually described with the Arrhenius plot, which has made it possible to determine that P. pastoris is a rather psychotrophic organism that can grow at a temperature as low as 12°C.49
Studies at different temperatures have indicated that methanol is more toxic at 30°C than TLFB processes, which use lower temperatures, although the latter uses much higher methanol concentrations. In addition, the AOX promoter presents a much higher transcriptional capacity.49 However, the environmental changes are perceived at other levels more than at transcript level; for example, proteins with modified catalytic properties or changes at metabolite level.51 All these data are an indicator that cultures at low temperatures in TLFB processes result in a reduced extracellular proteolysis for two reasons49: (a) purely thermodynamic reasons and (b) a reduced release of proteases from lysed cells. The proteolytic degradation of recombinant proteins secreted in high cell-density fermentations has been widely reported. In methylotrophic yeasts like Pichia the proteolytic activity and recombinant protein degradation are attributed to methanol metabolism combined with cell lysis at the end of fermentation. It has also been reported that proteolytic degradation can be reduced through the careful manipulation of pH and temperature.52 With regard to cell viability, both at very low (10°C) and very high temperatures (37°C), suboptimal growth conditions have been observed that prevent a fermentation process longer than 24h.51
Dragosits et al.6 determined that these reasons may not be the only ones to generate both cell density and low antibody production. They observed that the proteins involved in protein folding and secretion stayed constant or decreased during low growth temperatures (20°C and 25°C), whereas at 30°C, the transcript levels dropped, which would partly explain the low antibody production. This may be linked to other factors such as mRNA stability, protein synthesis rate and protein folding. All these effects may trigger a greater secretion capacity at 25°C and 20°C as a “secondary effect”. On the other hand, Zhong et al.53 observed that cultures at high temperatures presented a prolonged accumulation of the recombinant protein of interest in immature form in the endoplasmic reticulum, which led to high stress in this organelle and subsequent cell death. However, when the production rate of the recombinant protein fell as the culture temperature dropped, the stress of the endoplasmic reticulum was indeed mitigated, preserving the folding capacity of the endoplasmic reticulum and enhancing cell viability, which is consistent with other reports on the positive effects of cultures at low temperatures.
Innovation in industrial processes with an impact on efficient production is currently the major challenge for industry. A high number of enzymes are used at different levels of the process; the search for new alternatives with better characteristics has become a field of study of great interest.3Pichia pastoris, a methylotrophic yeast, is an established system for the production of heterologous proteins, particularly biopharmaceuticals and industrial enzymes.20 One of the most important factors to improve recombinant protein production, used nowadays to a large extent as biopharmaceuticals, involves understanding how the cell metabolism activates and mainly how this can be influenced under the different environmental conditions and/or medium culture to which the cells can be exposed. To maximize and optimize the production of recombinant products, recent molecular research has focused on numerous issues including the design of expression vectors, optimization of gene copy number, co-expression of secretory proteins such as chaperones, engineering of glycosylation and secretory pathways, etc. However, the physiological effects of different culture strategies are often difficult to separate from the molecular effects of the gene construct (e.g., cellular stress through over-expression or incorrect post-translational processing). Hence, overall system optimization is difficult, even though it is urgently required in order to describe and understand the behavior of new molecular constructs.20
ConclusionIt is important to consider that heterologous protein production depends of multiple factors (agitation, substrate concentration, inducer concentration, pH and others) that are difficult to control overall, because they all influence the protein expression system internally, and thus the quality and quantity of the desired product. Understanding the effects of carbon source and temperature on the metabolism of P. pastoris yeast is crucial to developing a suitable work methodology, starting with the choice of the appropriate promoter, as the use of a certain substrate and variation in the culture conditions can activate different metabolic pathways, activating enzymes that can help or harm the growth and development of the yeast and/or the recombinant protein production.
Conflicts of interestThe authors declare no conflicts of interest.
The authors acknowledge the financial support of the Comisión Nacional de Investigación Científica y Tecnológica de Chile (CONICYT N° 21110306) doctoral fellowship awarded to AZP (Chile), and 2012/50210-9 FAPESP (Sao Paulo Research Foundation, Brazil) (AP and JGF).