The aim of this study was to detect the IS900 region of Mycobacterium avium subsp. paratuberculosis (MAP) in bovine milk samples using real-time polymerase chain reaction (qPCR) and conventional PCR, and to study the agreement between these tests. A total of 121 bovine milk samples were collected from herds considered positive for MAP, from the State of Pernambuco, Brazil. MAP DNA was detected in 20 samples (16.5%) using conventional PCR and in 34 samples (28.1%) using qPCR. MAP DNA was detected in all of the 6 animal farms studied. Moderate agreement was found between qPCR and conventional PCR results, where the sensitivity and specificity of conventional PCR in relation to qPCR were 50% and 96.6%, respectively. Thus, the IS900 region of MAP was found in bovine milk samples from the State of Pernambuco. To the best of our knowledge, this is the first report of MAP DNA found in bovine milk in Northeast Brazil. We also demonstrated the qPCR technique is more sensitive than conventional PCR with respect to detection of MAP in milk samples.
Mycobacterium avium subsp. paratuberculosis (MAP) is a gram-positive, slow-growing bacillus of the family Mycobacteriaceae that possesses a cell wall rich in lipids, characteristic of this family. This microorganism is an intracellular pathogen, responsible for Johne's disease or paratuberculosis.1,2
Paratuberculosis has been previously investigated in several studies in Brazil.3 Its occurrence in Pernambuco has been described in dairy cattle based on clinical signs, histopathology, and results from enzyme-linked immune sorbent assay (ELISA) serology (32.3% positive samples), isolation (50% positive samples), and polymerase chain reaction (PCR) studies.4 Additionally, a prevalence rate of 2.7% (11/408) has been reported in the micro-region of Garanhuns, with 47.4% (9/19) outbreaks.5
The most common methods of MAP diagnosis in infected animals include isolation of bacteria from feces using selective culture media and antibody detection techniques such as ELISA. However, the greatest disadvantage of using culture media is the long incubation period, which can be as long as 16 weeks for a definitive diagnosis. ELISA can be performed within a few hours, although its sensitivity is estimated at only 45%, since antibodies to MAP may not be detectable in the initial stages of the infection.6,7
Molecular techniques to detect MAP, such as PCR, are rapid and qualitative in nature. Real-time PCR (qPCR) exhibits greater sensitivity than conventional PCR, and can determine the infective load in environmental samples, feces, milk, and cultures.7,8 One of the target genes used to detect MAP via PCR is the IS900 region, first described by Green et al.9 and independently identified by Collins et al.10 The discovery of the IS900 region of MAP enables the diagnosis of paratuberculosis even in the initial stages of infection. The specificity and sensitivity of PCR have been enhanced up to the point of detecting 1CFU of MAP in samples.11
MAP has been detected in several animal products and byproducts. A study conducted in Switzerland detected the presence of the IS900 region in 19.7% (273/1384) milk samples collected from milk storage tanks.12 A study in Cyprus reported 63 (28.6%) positive out of a total of 220 milk samples from tanks, using real-time PCR for IS900 and F57.13
The aim of this study was to detect the IS900 region of MAP in bovine milk samples from the State of Pernambuco (Brazil) using PCR and qPCR, and to investigate the agreement between these diagnostic tests.
Materials and methodsSamplingIn total, 121 bovine milk samples from the State of Pernambuco were collected from 6 dairy herds that already had a history of paratuberculosis. The animals were clinically healthy at the time of collection.
Sample collection and processingSample collectionThe cow teats were cleaned with water and disinfected with 70% alcohol prior to collection of milk samples. The first 3 jets of milk were discarded. Subsequently, approximately 50mL milk was pooled from the 4 mammary glands using sterilized and separate polypropylene tubes. The samples were stored in cool boxes containing recyclable ice and sent to the Laboratory of Bacteria in the Federal Rural University of Pernambuco for processing.
DNA extractionDNA extraction was performed using 2mL of each sample, which was centrifuged at 12,000×g for 10min, and the pellet was resuspended in 100μL buffered saline solution with sterile phosphate (pH 7.2). A commercial kita was used for extraction, following the manufacturer's instructions.
Polymerase chain reaction (PCR)The extracted DNA was amplified in a final volume of 15μL, containing the following: 5μL genomic DNA; 0.5μL each, of primers specific for IS900 at 20pM (DF: 5′-GACGACTCGACCGCTAATTG-3′ and DR-1: 5′-CCGTAACCGTCATTGTCCAG-3′); 2.75μL ultrapure Milli-Q water and 6.25μL PCR kit mixture,b as per the manufacturer instructions. The DNA was amplified in a thermocyclerc using the following conditions: initial denaturation at 96°C for 5min, followed by 35 cycles of denaturation at 95°C for 1min, annealing at 58°C for 1min and extension at 72°C for 3min, with a final extension at 72°C for 10min.14 The amplified product of 99bp, corresponding to MAP, was detected by electrophoresis in agarose gel (2%), followed by staining with blue green,d and visualization under ultraviolet light. Images of the bands were captured.
Polymerase chain reaction in real timeThe extracted DNA was amplified in a final volume of 25.0μL containing: 5μL genomic DNA; 1μL each, of primers specific for IS900 at 10pM (DF: 5′-GACGACTCGACCGCTAATTG-3′ and the DR-1: 5′-CCGTAACCGTCATTGTCCAG-3′), 5.5μL ultrapure Milli-Q water, and 12.5μL real-time PCR kit mixture,e according to the manufacturer instructions. The DNA was amplified in a thermocyclerf using the following conditions: initial denaturation at 95°C for 5min, followed by 40 cycles at 95°C for 20s and 60°C for 30s. The thermocycler software was used to monitor and interpret the results.
DNA from a MAP strain provided by the National Agricultural Laboratory in Minas Gerais identified as “Nakajima 1991” was used to standardize the qPCR. The melting curve consisted of 65°C for 90s for preparation, with a posterior gradual increase of 1°C every 5s from 60°C to 95°C. The peak of denaturation was at 82.9°C. The number of copies was determined as previously described by Rodrígues-Lázaro et al.15 The copy number detected was divided by 15, which corresponds to the average number of IS900 elements in the MAP genome.16 The DNA of the MAP strain was 10-fold serially diluted and used to obtain the standard curve ranging from 2.27×107 to 22.6 MAP cells. The efficiency of the primers was 95.0% and the coefficient of linear correlation (R2) was 0.99909, while the detection threshold (Ct) was between 9.58 and 30.08. The reactions were performed in duplicate. Finally, the number of copies in the positive field samples was quantified using the standard curve.
Sequencing of the samples and validation of conventional PCRIn order to validate the amplifications, one of the positives samples from the conventional PCR was purified using a commercial extraction kit,g as per the manufacturer instructions, before being sent for sequencing. Sequences with a Phred quality score above 20 were analyzed using the Basic Local Align Sequence Tool (BLAST; www.blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysisThe Kappa coefficient (K) was used to assess the agreement between tests. The qPCR was considered as a gold standard and the conventional interpretation of the K values adopted was as follows: 0.00–0.20=weak; 0.21–0.40=regular; 0.41–0.60=moderate; 0.61–0.80=good; and 0.81–1.00=very good agreement; negative values were interpreted as 0.00.17 Biostat softwareh was used to calculate the agreement, sensitivity, and specificity. The chi-squared test was used to determine associations between the tests, with a confidence interval of 95%.
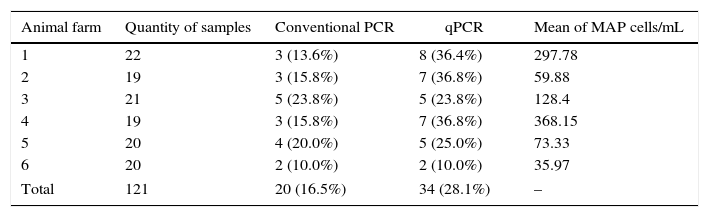
ResultsOf the 121 milk samples analyzed, the IS900 region of MAP was detected in 20 (16.5%) samples using conventional PCR and in 35 (28.1%) samples using qPCR. Milk samples contaminated with MAP were found in all six farms studied, ranging from 10.0 to 23.8% using conventional PCR and from 10.0 to 36.8% using qPCR (Table 1).
Frequency of positive milk samples by animal farm using conventional PCR and qPCR with the mean value of MAP cells/mL per animal farm determined by qPCR.
| Animal farm | Quantity of samples | Conventional PCR | qPCR | Mean of MAP cells/mL |
|---|---|---|---|---|
| 1 | 22 | 3 (13.6%) | 8 (36.4%) | 297.78 |
| 2 | 19 | 3 (15.8%) | 7 (36.8%) | 59.88 |
| 3 | 21 | 5 (23.8%) | 5 (23.8%) | 128.4 |
| 4 | 19 | 3 (15.8%) | 7 (36.8%) | 368.15 |
| 5 | 20 | 4 (20.0%) | 5 (25.0%) | 73.33 |
| 6 | 20 | 2 (10.0%) | 2 (10.0%) | 35.97 |
| Total | 121 | 20 (16.5%) | 34 (28.1%) | – |
The number of DNA copies, determined by qPCR in all positive samples exhibited a mean value of 160.59MAP cells/mL, with minimum and maximum values of 12.2 and 1126.66 MAPcells/mL, respectively. The mean value of the quantifications per animal farm for the positive samples ranged from 35.97 to 368.15MAP cells/mL (Table 1).
The agreement between conventional PCR and qPCR, analyzed by the Kappa coefficient, was 0.53, which is moderate (χ2=38.08; p<0.000). The values for sensitivity and specificity of the conventional PCR were 50% and 96.6%, respectively.
The sequencing of the field sample exhibited a similarity of 98% with the DNA of M. avium subsp. paratuberculosis (MAP4, complete genome) deposited at the National Center for Biotechnology Information.
DiscussionThis is the first record of the detection of MAP in bovine milk in Northeast Brazil. The presence of MAP in milk has been previously reported in Brazil by Carvalho et al.,18 who found the IS900 region of DNA in 8/222 (3.6%) samples of raw bovine milk from Viçosa, Minas Gerais.
MAP has been detected previously in bovine milk in other countries. In Switzerland, DNA of IS900 was found in 273 (19.7%) of 1384 samples from milk tanks.12 Slana et al.13 collected a total of 220 milk samples from tanks in Cyprus and found 63 (28.6%) positive samples (IS900 and F57) using real-time PCR.
Paratuberculosis is considered uncommon in Brazil; however, outbreaks of the disease have been reported in 11 states of the Federation.3,19 In Pernambuco, Mota et al.4 reported the occurrence of paratuberculosis in a bovine dairy herd by isolating MAP from feces samples, with genetic confirmation of the IS900 region using PCR.
The high frequency of positive milk samples found in this study could be explained by the fact that the milk samples were collected from herds considered positive for paratuberculosis. It is important to note that the actual prevalence rate could be even higher, since not all infected animals eliminate the bacterium via milk.12
Based on the health risks due to MAP and considering that this microorganism is implicated in Crohn's disease in humans, rapid and sensitive tests for detection of this bacterium in milk are urgently required.7 In this study, the number of milk samples found positive via qPCR (34 samples) was higher than that via conventional PCR (20 samples). The qPCR technique is more efficient in detecting pathogenic bacteria and has several advantages over conventional PCR, including the potential to quantify the bacteria, the minimal risk of cross-contamination, as well as greater sensitivity and specificity that can be enhanced by the use of probes.20
Sonawane and Tripathi21 reported that 2 groups of tissues (multibacillary and paucibacillary) were used to perform conventional PCR to detect the IS900 region of MAP. They recorded sensitivity levels of 82.6% and 13.3% for the multibacillary and paucibacillary groups, respectively. In the same study, qPCR recorded sensitivity levels of 100% in the detection of MAP for both groups.
Upon analysis of the agreement between nested PCR and qPCR in terms of detecting MAP, Fang et al.22 obtained a Kappa index of 0.85. This value, which is higher than that found in the present study, could be explained by the fact that nested PCR is more sensitive than conventional PCR, which was used in the present study for comparison with qPCR.23
In the present study, 3 samples tested positive using conventional PCR and not by qPCR. The reason for this is unclear; however, a non-homogenous distribution and/or small quantity of MAP bacteria in the sample could be responsible for the result.22 Another possible reason is the presence of some inhibitors, such as calcium, in the extracts, which even in small quantities, could inhibit Taq and/or compete with magnesium, thus reducing the efficiency of qPCR.24 This, however, does not refute the finding that the qPCR method reliably detected the presence of MAP DNA in a greater proportion when compared with conventional PCR.
Direct detection of viable MAP using cultures is considered the gold standard. However, MAP can be detected more quickly and with approximately 10 times higher sensitivity using qPCR.13,25 This discrepancy between the methods could be explained by the low concentration of MAP in the sample, since qPCR is a more sensitive detection technique than culturing, from both, feces and milk.26
Considering that bovines with sub-clinical infections can eliminate very low quantities of MAP via milk,27 it is essential to use adequate and sensitive diagnostic tests to detect this bacterium. The sensitivity of qPCR, in terms of detecting MAP in milk samples, has been reported to range from 1 to 100CFU/mL milk.28 Additionally, determination of MAP bacterial load in milk samples is also possible with qPCR. Conventional PCR as well as culture methods are not quantitative and culture methods also cannot assess this factor due to the decontamination process of the samples, which decreases the quantity of MAP in the sample.7
Raw bovine milk has received considerable attention, as it is known to be an important factor in the transmission of MAP from cow to calf. A study has shown that milk and the colostrum may be risk factors for transmission of MAP infection.29
MAP still remains to be implicated in Crohn's disease, and if humans can be infected by MAP, there are several forms of transmission, including transmission between humans. However, since animals are the main source of these bacteria, transmission from animals to humans is more likely,30 which can occur via food. Viable MAP have been recovered from pasteurized milk samples in Brazil31 and other countries as well.32 The presence of MAP in milk products such as cheese has been confirmed in Greece and the Czech Republic.33
ConclusionIn this study, the IS900 region of MAP was detected in bovine milk samples obtained from the State of Pernambuco, and to our knowledge, this is the first such report of MAP DNA in bovine milk in Northeast Brazil. We also demonstrated that the qPCR technique is more sensitive than conventional PCR in terms of detecting MAP in milk samples.
Ethical approvalThe Ethics Committee on Animal Use of the Federal Rural University of Pernambuco approved the ethical procedures of this study under license No. 034/2013.
Conflicts of interestThe authors declare no potential conflicts of interest with respect to this study or the authorship and/or publication of this article.
DNA Easy Blood and Tissues Kit®, Qiagen Biotechnology Brazil Ltda., São Paulo, Brazil.