The severity of Helicobacter pylori-related disease is correlated with the presence and integrity of a cag pathogenicity island (cagPAI). cagPAI genotype may have a modifying effect on the pathogenic potential of the infecting strain. After analyzing the sequences of cagPAI genes, some strains with the East Asian-type cagPAI genes were selected for further analysis to examine the association between the diversity of the cagPAI genes and the virulence of H. pylori. The results showed that gastric mucosal inflammatory cell infiltration was significantly higher in patients with East Asian-type cagPAI genes H. pylori strain compared with mosaicism cagPAI genes H. pylori strain (p<0.05). H. pylori strains with the East Asian-type cagPAI genes were closely associated with IL-8 secretion in vitro and in vivo compared with H. pylori strains with the mosaicism cagPAI genes (p<0.01). H. pylori strains with East Asian-type cagPAI genes are able to strongly translocate CagA to host cells. These results suggest that H. pylori strains with East Asian-type cagPAI genes are more virulent than the strains of cagPAI gene/genes that are Western type.
Helicobacter pylori (H. pylori) is a gram-negative bacterium and is the main pathogenic factor of gastroduodenal diseases. H. pylori contains many virulence factors, including the cag pathogenicity island (cagPAI), that are involved in the pathogenesis of several diseases.1,2H. pylori cagPAI consists of approximately 30 genes encoding for proteins of the type IV secretion system (T4SS), which transfers CagA and peptidoglycan into host epithelial cells, resulting in increased cellular release of interleukin 8 (IL-8).3,4 Studies have also demonstrated that H. pylori strains with a functional T4SS are more frequently associated with the etiology of H. pylori.5,6
H. pylori is a highly genetically diverse species that is attributed to its high rate of DNA recombination.7–9 It has been reported in Japan that the genetic diversity within the cagPAI may have modifying effects on the pathogenic potential of infecting strains.10 The similarity coefficient revealed that all cagPAI genes could be placed into two major groups, East Asian-type or Western-type. Most of cagPAI genes belong to the East Asian-type and is found associated with high risk of developing gastric cancer in some regions. Noticeably, in some regions, some strains of the Western-type of several cagPAI gene/genes are of East Asian-type of other cagPAI genes, potentially due to the mosaicism of cagPAI genes in H. pylori and genetic recombination.11
Much work has been reported on the genetic diversity of cagPAI, particularly on the genotype of cagA.12,13 Minimal research has been conducted in applying cagPAI genotype analysis to evaluate the virulence of H. pylori. In some H. pylori strains, the two parts of the cagPAI genes are interposed by a segment called insertion sequence 605 (IS605): the upstream as cagII region, and the downstream as cag I region. In this study, some cag PAI genes in cag II (cagT, cagX) and cagI region (cagI, cagL, cagM) were selected for sequencing. The virulence of the strains with East Asian-type cagPAI genes was evaluated by comparing strains with mosaic cagPAI genes.
Materials and methodsPatients and samplingPatients with abdominal symptoms were clinically examined, and biopsy samples were taken for H. pylori isolation and histological analysis in Weihai Municipal Hospital (affiliated with Dalian Medical University) from June 2010 to June 2014. None of the patients underwent antimicrobial therapy, or have taken proton pump inhibitors (PPIs) or non-steroidal anti-inflammatory drugs a month before their inclusion in the study. Written and informed consent was obtained from all patients, and the study was conducted upon approval by the ad hoc Ethical Committee of Weihai Municipal Hospital.
H. pylori cultivation and identificationBiopsy specimens were collected in brain heart infusion broth (Oxoid, United Kingdom), and dispersed by using the tissue homogenizer. Every homogenate was inoculated onto Campylobacter agar (Oxoid, United Kingdom) with 8% sheep blood and H. pylori selective supplement (Oxoid, United Kingdom) under microaerophilic condition (5% O2, 10% CO2 and 85% N2) at 37°C for 72h. Small dew drop colonies of H. pylori were selected for further phenotypic analysis by PCR of the 16SrRNA gene sequence as previously described.11
Sequence analysis of cagPAI genesThe presence of five cagPAI genes, which spread over the cag I (cagI, cagL, cagM) and cag II regions (cagT, cagX) in H. pylori strains, was analyzed by PCR using five sets of primers listed in Table 1. The amplicons were selected for sequence analysis by Life Technologies Corporation. After the full-length amino acid sequences of each gene were translated from the nucleotide sequences by Primer 5.0, the phylogenetic trees of CagPAI proteins were constructed by the neighbor-joining method of Saitou and Nei by using a program called MEGA. All cagPAI genes in the isolates could be placed into two major groups. Most H. pylori strains from the region are either East Asian-type or Western-type. Three previously reported Western strains have been identified so far: 26695 (UK), J99 (USA), and NCTC11637 (Australia).
Polymerase chain reaction (PCR) primer pairs used for cagPAI genes in Helicobacter pylori isolates.
| Region | Primer | Sequence | Amplicon size (bp) |
|---|---|---|---|
| cagI | f: | ATAGAATTCACAGAAGTAGTAATAACGCTTGAAC | 1086 |
| r: | AGACTCGAGTTTGACAATAACTTTAGAGCTAG | ||
| cagL | f: | GATGGATCCGAAGATATAACAAGCGGTTT | 654 |
| r: | GCCCTCGAGTTTAACAATGATCTTACTTGA | ||
| cagM | f: | TAGGGATCCGAGCAGTTTGGTTCATTT | 1149 |
| r: | CGCCTCGAGCTATTCAAAGGGATTATTC | ||
| cagT | f: | TCCGGATCCATGAAAGTGAGAGCAAGTGT | 843 |
| r: | GCCAAGCTTTCACTTACCACTGAGCAAAC | ||
| cagX | f: | GGAATTCATGGGGCAGGCATTTTTTA | 1569 |
| r: | GGGTCGACTTATTTATCTCTGACAAGAGGGAG | ||
| cagA conservative region | f: | GATAGGGATAACAGGCAAGC | 297 |
| r: | GGGGGTTGTATGATATTTTC | ||
| cagA 3′ variable region | cag2 | GGAACCCTAGTCGGTAATG | Uncertain |
In the present study, the strains were classified based on the genotypes of cagPAI genes: East Asian-type cagPAI genes group (all five cagPAI genes), and mosaicism cagPAI genes group (less than five cagPAI genes identified as East Asian-type, and containing more than one Western-type cagPAI gene except cagA).
Analysis of cagA status and 3′ variable region of cagAPCR analyses were performed to analyze cagA status and cagA 3′ variable region using specific primers (Table 1). After PCR, the amplified PCR products were electrophoresed in 2% agarose gels and examined under UV illumination. The amplicons of cagA 3′ variable region were sequenced by Life Technologies Corporation.
Histological analysisStomach biopsy specimens from each patient were examined by an experienced pathologist. For each biopsy specimen, the grades of inflammatory cell infiltration, gastric mucosal atrophy, and H. pylori density were scored on the basis of the updated Sydney System (0, none; 1, mild; 2, moderate; 3, severe).14
Mucosal IL-8 level analysis in biopsy samplesIL-8 levels in the biopsy specimens were measured after homogenization using an enzyme-linked immunosorbent assay (ELISA) (Invitrogen). Briefly, the supernatants from homogenized specimens were obtained by centrifugation (10,000×g for 15min), and the total proteins of the supernatants were measured by using the Bradford assay (Bio-Rad, Richmond, CA). IL-8 levels in samples were expressed as pg/mg of protein.
Determination of IL-8 secretion from GES-1 cells co-cultured with H. pylori strainsThe strains were selected and co-cultured with GES-1 cells for analysis of IL-8 secretion. Briefly, GES-1 cells were seeded at a density of 8×104cells/well in 96-well plates. H. pylori was harvested from agar dishes and washed twice with PBS before being added to culture wells at a MOI of 100. The cell culture media were collected at 24h. IL-8 levels were detected by ELISA (Invitrogen) according to the manufacturer's protocol. The experiments were performed twice independently.
CagA Translocation AssayGES-1 cells infected with H. pylori (1:100) as described above were washed with PBS until no H. pylori were found adhered to cells as observed under a microscope. Cells were then scraped and centrifuged at 4000rpm for 30min. Cells were suspended in RIPA Lysis Buffer on ice for 30min. After centrifuging at 13,500rpm for 30min, the supernatant was collected to analyze CagA by Western blotting. Colony-forming units (CFU) assay was performed to confirm that no H. pylori were detected on scraped cells.
Statistical analysisDifferences between the Asian-type strains group and the mosaic-type strains group were analyzed using Student's t test and Chi-square test. p values less than 0.05 was considered significant.
ResultsAfter sequence analysis of cagPAI genes (Fig. 1), 76 strains with East Asian-type cagPAI genes were isolated from gastroduodenal diseases, including chronic gastritis (CG, n=29), gastric ulcer (GU, n=21), duodenal ulcer (DU, n=19), and gastric cancer (GC, n=7). Twenty-six strains with mosaicism cagPAI genes were isolated from chronic gastritis (CG, n=14), gastric ulcer (GU, n=6), duodenal ulcer (DU, n=4), and gastric cancer (GC, n=2). Isolates with East Asian-type cagPAI genes were found at lower frequency in CG (38.2%, 29/76) than those with mosaicism of cagPAI genes in CG (53.8%, 14/26, p<0.01).
PCR products were obtained from all isolates of H. pylori. After nucleotide analysis, cagA 3′ variable region were grouped as East Asian-type or Western-type. The East Asian-type (521bp) possessed three EPIYA motifs, while the Western-type (502bp) possessed two EPIYA motifs and one EPIYT (Fig. 2). All 76 strains with East Asian-type cagPAI genes harbored East Asian-type, and only 2 strains possessed Western-type in 26 strains with mosaicism cagPAI genes.
Histological analysis of H. pylori density, inflammatory cell infiltration, and atrophy in the biopsy specimensFurther histopathologic evaluations of biopsy specimens were performed on 42 patients with East Asian-type cagPAI genes H. pylori strains and 26 patients with mosaicism cagPAI genes H. pylori strains. The biopsy specimens of East Asian-type cagPAI genes group were selected randomly for histological analysis.
Gastric mucosal inflammatory cell infiltration was significantly higher in East Asian-type cagPAI genes group than mosaicism cagPAI genes group (p<0.05) (Fig. 3). However, there were no significant differences in the grade of H. pylori density and gastric mucosal atrophy according to the diversity of cagPAI genes (Table 2). This suggested that the status of cagPAI genotype is associated with progression of gastric mucosal inflammatory cell infiltration.
Histological differences among the two cagPAI groups.
| Parameter | East Asian-type cagPAI genes group (n=42) | Mosaicism cagPAI genes group (n=26) |
|---|---|---|
| H. pylori density | 1.52±0.41 | 1.63±0.52 |
| Inflammatory cell infiltration | 1.71±0.68a | 1.03±0.42 |
| Atrophy | 0.51±0.17 | 0.47±0.16 |
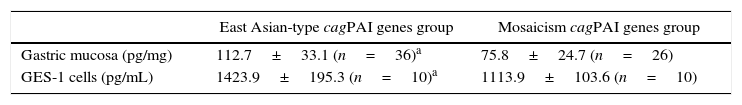
We randomly selected 36 patients with East Asian-type cagPAI genes H. pylori strains and 26 patients with mosaicism cagPAI genes H. pylori strains to measure gastric mucosal IL-8 levels. The gastric mucosal IL-8 level in patients with East Asian-type cagPAI genes H. pylori strains was significantly higher (112.7±33.1pg/mg) than those containing mosaicism cagPAI genes H. pylori strains (75.8±24.7pg/mg, p<0.01).
IL-8 production in GES-1 cells that were co-cultured with 20 H. pylori strains was also examined. These included 10 strains with East Asian-type cagPAI genes and 10 strains with mosaicism cagPAI genes. IL-8 production (1423.9±195.3pg/mL) was significantly higher in strains with East Asian-type cagPAI genes than in those with mosaicism cagPAI genes (1113.9±103.6pg/mL, p<0.01) (Table 3). These results suggested that the cagPAI genotype of H. pylori strain is important for inducing IL-8 production in vivo and in vitro.
CagA delivery to host cellsTen H. pylori strains with East Asian-type cagPAI genes and 10 strains with mosaicism cagPAI genes were randomly selected for further analysis of CagA translocation. After infection of GES-1 cells by H. pylori, cells with translocated CagA were found more increased in East Asian-type cagPAI genes group than mosaicism cagPAI genes group. These results indicated that the translocation of CagA to host cells was less effective in strains with mosaicism cagPAI genes (Fig. 4).
DiscussionH. pylori cagPAI is a major virulence determinant in H. pylori-related diseases. In comparison with cagPAI-negative strains, infection with cagPAI-positive H. pylori strains significantly increases the risk of developing severe gastric mucosal inflammation, duodenal ulcers, and gastric cancers.15,16 The cagA gene is part of cagPAI, and CagA is the primary virulence factor of H. pylori that sufficiently induces tumorigenesis in cell and animal models. CagA has also been reported to be epidemiologically associated with higher risk of developing gastric cancer.17,18 Based on the sequence of the 3′ region, cagA was classified into 2 types: East Asian-type and Western-type. Intriguingly, both in vitro and in vivo studies have clearly showed that East Asian-type CagA protein is more carcinogenic than Western-type CagA protein.19 In this region, most strains possess East Asian-type of cagA with three EPIYA motifs.
In a littoral region of Northeast China, nearly 100% of the strains possess cagA, and most isolates of cagA were included in East Asian-type.20 Noticeably, a high incidence of atrophic gastritis and gastric cancer has also been reported in this region.21 Early studies showed that the distinct distribution of cag PAI diversity in this region may be involved in the development of atrophic gastritis, potentially increasing the risk of worsened outcome in different diseases based on phylogenetic analysis of cagPAI genes.11 We hypothesize that the T4SS encoded by Eastern-type cagPAI genes in the region results in the ability of H. pylori to increase the risk of developing gastric cancer.
In the study, isolates with East Asian-type cagPAI genes were found at lower frequency in CG, compared to strains with mosaicism cagPAI genes in CG. Two cagPAI genotypes of H. pylori strains were selected based on the phylogenetic analysis of full-length CagPAI proteins, including East Asian-type cagPAI and mosaicism cagPAI genes strains. Some strains were selected to study their virulence, and to examine whether there is an association between the diversity of the cagPAI genes and the virulence of H. pylori. We found that gastric mucosal inflammatory cell infiltration was significantly higher in patients with East Asian-type cagPAI genes H. pylori than others with mosaicism cagPAI genes strains. These observations suggest that cagPAI genotype is associated with progression of gastric mucosal inflammatory cell infiltration. Meanwhile, we also found that H. pylori strains with the East Asian-type cagPAI genes are closely associated with IL-8 secretion compared with H. pylori strains with the mosaicism cagPAI genes. Intriguingly, H. pylori strains with the East Asian-type cagPAI genes can easily translocate CagA to host cells.
H. pylori exhibits extensive genetic diversity and rapid allelic diversification attributed to its high mutation rate and frequent recombination in different diseases.22–24 We believe that the mosaicism in cagPAI genes is best explained by genetic recombination. Thus, the genetic recombination in cagPAI between the west strain and East strain could increase the diversification and virulence of H. pylori.
In conclusion, H. pylori with the East Asian-type cagPAI genes is associated with gastric mucosal inflammatory cell infiltration and IL-8 secretion, which seems to be more virulent compared with other strains.
Conflicts of interestThe authors declare that they have no conflicts of interest concerning this article.
This work was supported by Shandong Provincial Natural Science Foundation, China (No. ZR2015HM072) and Jiangsu Key Laboratory of Medical Science and Laboratory Medicine (No. JSKLM-2014-014).