Presence of extended spectrum beta-lactamases (ESBL) in bacteria is a growing health concern of global significance. The local, regional, national, and international epidemiological studies for extended spectrum beta-lactamases-producing Enterobacteriaceae and their encoding genes in foods are still incomplete. The objective of this study was to determine the occurrence of extended spectrum beta-lactamases-producing Enterobacteriaceae and the characteristics of their encoding genes from a total of 250 samples of various foods of animal-origin (100 raw chicken meat, 100 raw cow milk, and 50 raw cow milk cheese) sold in Turkey. Overall, 55 isolates were positive as extended spectrum beta-lactamases-producing Enterobacteriaceae. The most prevalent extended spectrum beta-lactamases-producing strain were identified as Escherichia coli (80%), followed by Enterobacter cloacae (9.1%), Citrobacter braakii (5.5%), Klebsiella pneumoniae (3.6%), and Citrobacter werkmanii (1.8%) by Vitek® MS. The simultaneous production of extended spectrum beta-lactamases and AmpC was detected in five isolates (9.1%) in E. coli (80%) and E. cloacae (20%). The frequency rates of blaTEM, blaCTX-M, and blaSHV were 96.4%, 53.7%, and 34.5%, respectively. The co-existence of bla-genes was observed in 82% of extended spectrum beta-lactamases producers with a distribution of blaTEM & blaCTX-M (52.7%), blaTEM & blaSHV (20%), blaTEM & blaCTX-M & blaSHV (12.7%), and blaSHV & blaCTX-M (1.8%). The most prevalent variant of blaCTX-M clusters was defined as blaCTX-M-1 (97.2%), followed by blaCTX-M-8 (2.8%). In summary, the analysed foods were found to be posing a health risk for Turkish consumers due to contamination by Enterobacteriaceae with a diversity of extended spectrum beta-lactamases encoding genes.
Antibiotics are used as veterinary and human medicines for treatment, control and prevention of infectious diseases. However, their repeated off-label over use can have un-anticipated adverse effects including development of antibiotic resistance in the bacteria towards modern beta-lactam antibiotics.1
One of the antibiotic resistance mechanisms in the bacteria is the production of specific enzymes such as beta-lactamase to break down the four-atom beta-lactam ring of the antibiotics. When this ring is opened through hydrolysis by these enzymes, the antimicrobial properties of the antibiotics are completely lost.2 The extended spectrum beta-lactamases (ESBL) are the rapidly evolving plasmid-mediated group of beta-lactamases.3 ESBL can hydrolyse penicillins, first, second and third-generation cephalosporins, and aztreonam (but not cephamycins or carbapenems). The activity of ESBL can be inhibited by beta-lactamase inhibitors such as clavulanic acid (CLA).4 The antibiotic resistance leads to increased morbidity, mortality and the cost of treating infections, in particular, those caused by ESBL-producing bacteria.5 The most prevalent extended spectrum beta-lactamases are TEM, SHV, and CTX-M variants.6 The CTX-M variants are also clustered in five major groups: 1, 2, 8, 9, and 25.7 After 2000s, CTX-M type enzymes have exhibited a rapid growth over a wide range geographic areas, and have become much more prevalent than TEM and SHV type enzymes.8
The ESBLs are mainly encoded by plasmids and mobile genetic elements such as integrons, insertion sequences, transposons and plasmids. These genetic elements are easily transferable to other bacteria such as those belonging to Enterobacteriaceae.5,9 The widespread use of third generation cephalosporins and aztreonam has led to the emergence and dissemination of ESBL producing strains and their encoding genes, in particular, among Enterobacteriaceae associated with severe enteric illnesses.10–13
The major genes responsible for ESBL production include TEM genes (blaTEM), SHV genes (blaSHV), and CTX-M genes (blaCTX-M).14 This continued evolution is a serious threat to the public health by limiting the ability to treat bacterial infections.15,16 Most of the medicines under human consumption are antibiotics. Moreover, antibiotic use in veterinary medicine and for growth promotion and disease prevention in agriculture, aquaculture and horticulture is also a huge problem of antibiotic resistance, since most of the antibiotics manufactured (100,000–200,000 tonnes per year) is estimated to be massively consumed by these sectors, especially in the food-producing animals for last 60 years.17,18 Therefore, food-producing animals and foods of animal-origin are under suspicion for being transmission vectors for colonisation and infection of the humans with ESBL-producing Enterobacteriaceae.11,16
The potential contribution of foods to human health risks by dissemination of plasmid-born ESBLs should be epidemiologically assessed because local, regional, national and international information related to this emerging phenomenon is largely incomplete in various geographical locations, including Turkey.9,19 The Turkish authorities accept that the use of antibiotics in the food animals to make them grow faster and/or to prevent disease cannot be controlled effectively due to the economic benefits of food-animals, as the sector largely ignores risks associated with human and animal health.20 Therefore, chicken meat, raw milk and cheese can be identified as high risk-carrying contributors for ESBL-producing Enterobacteriaceae and their encoding genes.9
The objective of this study was to determine the occurrence of ESBL-producing Enterobacteriaceae and the frequency rates of corresponding encoding genes in various foods of animal origin sold in Turkey.
Materials and methodsReference culturesAn ESBL-negative strain Escherichia coli ATCC®25922 and an ESBL-positive strain Klebsiella pneumoniae ATCC®700603 were used as negative and positive controls, respectively, in the phenotypic and genotypic methods.
Sampling, sample preparation, isolation and identification of ESBL suspicious bacteriaDuring the period of May 2014 to November 2014, a total of 250 food samples of animal origin (100 packed raw chicken meat from public bazaars, markets and poultry farms, 100 raw cow milk from containers and holding tanks in bazaars and dairy farms, and 50 unpacked raw cow milk cheese from bazaars and markets) were randomly collected in different cities of Marmara, Aegean and Black Sea regions. All samples were placed into sampling bags, and taken to the laboratory in a thermobox container at 4°C. The sample preparation and further examinations were quickly started after the sampling.
Ten millilitres of raw milk in 90mL of Enterobacteriaceae Enrichment Broth (LABM, Lancashire, England), and 25g either of raw cow milk, cheese or raw chicken meat in 225mL of the mentioned broth was homogenised in a sterile bag (Interscience, Saint Nom, France) for 2min by a stomacher (EasyMix, AES Chemunex, Rennes, France), and followed by an aerobic incubation at 37°C for 18–24h.21 After that, 10μL of the pre-enriched suspension was directly streaked onto Chromatic™ ESBL selective media (Liofilchem, Istanbul, Turkey). The inoculated plates were incubated at 37°C for 18–24h under aerobic condition as per to the manufacturer's instructions. The presumptive ESBL colonies were then sub-cultured onto Tryptic Soy Agar (LABM, Lancashire, England), followed by an incubation at 37°C for 18–48h. The species identification of suspected ESBL isolates was performed by Vitek® MS (bioMérieux, Marcy l’Etoile, France).
Screening and confirmation of ESBLsPresumptive ESBL-producers were screened using cefpodoxime (CPD; 10μg), cefotaxime (CTX; 30μg), and ceftazidime (CAZ; 30μg) antibiotic discs (MAST Group, UK). A 0.5 McFarland standard inoculum of the isolate was spread onto Mueller Hinton Agar (LABM, Lancashire, England) plates. The discs were inserted on the plate, and allowed for incubation at 37°C for 18–24h. The breakpoints with zone diameters were evaluated according to the criteria established by.22 The ESBL Set D67C (MAST Group, UK) with a combination of CPD, CTX, and CAZ±clavulanate (CLA, 10μg) was used for disc diffusion confirmation test. The disc inserted plates were incubated at 37°C for 18–24h. The zone of inhibition was evaluated according to the criteria described by 22.
Determination of minimal inhibitory concentration (MIC)Micronaut-S beta-lactamase VII Plate (Merlin Diagnostika, Berlin, Germany) was used for the phenotypic detection of ESBL-production, including AmpC, metallo-beta-lactamase (MBL), and carbapenemase (KPC) according to the criteria described by 22. A 50μL aliquot of 0.5 McFarland-standardised microbial suspension of the isolate was initially vortexed in 10mL of Mueller Hinton Broth (Merck, Darmstadt, Germany). Subsequently, 100μL of this suspension was pipetted into each well of the plate. After that, the plate was incubated overnight at 37°C. The reading was recorded using a Thermofisher Multiskan FC Spectrometer. The MIC analysis was automatically performed by the MCN6 Software (Sifin, Berlin, Germany).
Extraction of bacterial DNATotal DNA of ESBL-producing isolates was extracted using the GENESpin DNA Isolation Kit (Eurofins, Hamburg, Germany) according to the manufacturer's instructions. The extracted DNA samples were stored at –20°C for further molecular analyses.
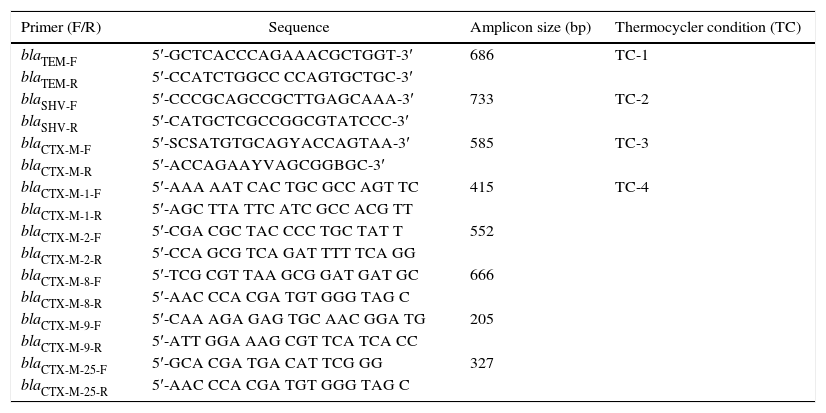
Characterisation of the bla variantsThe primer pairs as previously described by 23 were used for screening of blaTEM, blaSHV and blaCTX-M. The blaCTX-M variants (blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, and blaCTX-M-25) were sub-characterised according to 24. All primers were prepared by Fullgen Biotechnology and İontek (İstanbul, Turkey). The amplification was performed by a Thermal Cycler PTC-0200G DNA Engine (BioRad, Istanbul, Turkey). The blaTEM, blaSHV and blaCTX-M were amplified in a singleplex PCR assay while a multiplex PCR assay was performed for blaCTX-M variants. The PCR mixtures were prepared in a final volume of 25μL. All PCR conditions were optimised again as shown in Table 1. Gel-electrophoresis of the PCR products was performed on a 1.5% agarose-gel. The amplicons were photographed by GelDoc 2000 imaging system (BioRad, Istanbul, Turkey). Finally, the analysis was carried-out using Quantity One 4.6.3 GelDoc XR Software (BioRad, Istanbul, Turkey).
Primer design, sequences, amplicon sizes and thermocycler conditions for bla-genes according to 23, 24.
| Primer (F/R) | Sequence | Amplicon size (bp) | Thermocycler condition (TC) |
|---|---|---|---|
| blaTEM-F | 5′-GCTCACCCAGAAACGCTGGT-3′ | 686 | TC-1 |
| blaTEM-R | 5′-CCATCTGGCC CCAGTGCTGC-3′ | ||
| blaSHV-F | 5′-CCCGCAGCCGCTTGAGCAAA-3′ | 733 | TC-2 |
| blaSHV-R | 5′-CATGCTCGCCGGCGTATCCC-3′ | ||
| blaCTX-M-F | 5′-SCSATGTGCAGYACCAGTAA-3′ | 585 | TC-3 |
| blaCTX-M-R | 5′-ACCAGAAYVAGCGGBGC-3′ | ||
| blaCTX-M-1-F | 5′-AAA AAT CAC TGC GCC AGT TC | 415 | TC-4 |
| blaCTX-M-1-R | 5′-AGC TTA TTC ATC GCC ACG TT | ||
| blaCTX-M-2-F | 5′-CGA CGC TAC CCC TGC TAT T | 552 | |
| blaCTX-M-2-R | 5′-CCA GCG TCA GAT TTT TCA GG | ||
| blaCTX-M-8-F | 5′-TCG CGT TAA GCG GAT GAT GC | 666 | |
| blaCTX-M-8-R | 5′-AAC CCA CGA TGT GGG TAG C | ||
| blaCTX-M-9-F | 5′-CAA AGA GAG TGC AAC GGA TG | 205 | |
| blaCTX-M-9-R | 5′-ATT GGA AAG CGT TCA TCA CC | ||
| blaCTX-M-25-F | 5′-GCA CGA TGA CAT TCG GG | 327 | |
| blaCTX-M-25-R | 5′-AAC CCA CGA TGT GGG TAG C |
TC-1: 1 cycle for 15min at 95°C, followed by 35 cycles for 1min at 95°C, 1min at 63°C, and 1min at 72°C, and finally 10min at 72°C.
TC-2: 1 cycle for 15min at 95°C, followed by 30 cycles for 30s at 94°C, 30s at 58.5°C, and 45s at 72°C, and finally 10min at 72°C.
TC-3: 1 cycle for 15min at 95°C, followed by 30 cycles for 30s at 94°C, 30s at 55°C, and 45s at 72°C, and a final elongation at 72°C for 10min.
TC-4: 1 cycle for 15min at 94°C; 5 cycles for 25s at 94°C, 40s at 52°C, and 50s at 72°C, followed by 22 cycles for 25s at 94°C, 40s at 54°C, and 50s at 72°C, and a final elongation at 72°C for 6min.
The Kruskal–Wallis H-test was used for multiple comparison of the proportions of ESBL producers in their sampling groups. The test statistic H was approximated by a χ2 distribution. All statistical analyses were performed using SPSS 19 (SPSS Inc., USA) statistical package programme, and a P-value<0.05 was considered as statistically significant.
ResultsIn this study, we screened, for ESBL-producing Enterobacteriaceae, 100 raw chicken meat samples, 100 raw cow milk samples, and 50 raw cow milk cheese samples from different cities of Marmara, Agean and Black Sea regions, and subsequently characterised their encoding blaTEM, blaSHV and blaCTX-M variants.
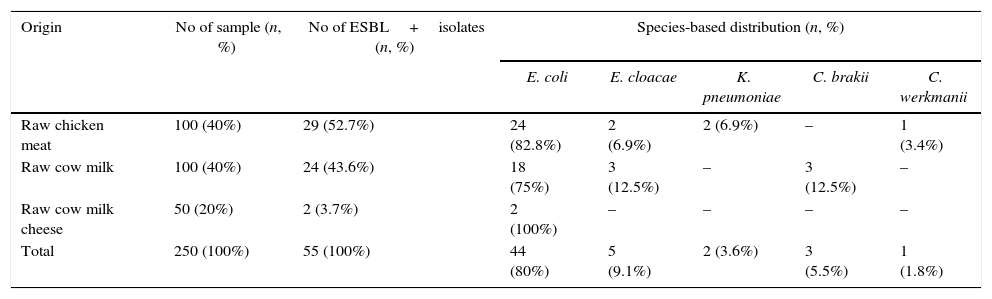
Phenotypic resultsThe phenotypic testing indicated that a total of 55 isolates (29 in raw chicken meat, 24 in raw cow milk, and 2 in raw cow milk cheese) from 250 food samples of animal origin were positive for ESBL-producing Enterobacteriaceae. The most prevalent phenotype was E. coli (80%), followed by Enterobacter cloacae (9.1%), Citrobacter braakii (5.5%), K. pneumoniae (3.6%), and Citrobacter werkmanii (1.8%) (Table 2).
Screening results of the samples for ESBL-producing Enterobacteriaceae.
| Origin | No of sample (n, %) | No of ESBL+isolates (n, %) | Species-based distribution (n, %) | ||||
|---|---|---|---|---|---|---|---|
| E. coli | E. cloacae | K. pneumoniae | C. brakii | C. werkmanii | |||
| Raw chicken meat | 100 (40%) | 29 (52.7%) | 24 (82.8%) | 2 (6.9%) | 2 (6.9%) | – | 1 (3.4%) |
| Raw cow milk | 100 (40%) | 24 (43.6%) | 18 (75%) | 3 (12.5%) | – | 3 (12.5%) | – |
| Raw cow milk cheese | 50 (20%) | 2 (3.7%) | 2 (100%) | – | – | – | – |
| Total | 250 (100%) | 55 (100%) | 44 (80%) | 5 (9.1%) | 2 (3.6%) | 3 (5.5%) | 1 (1.8%) |
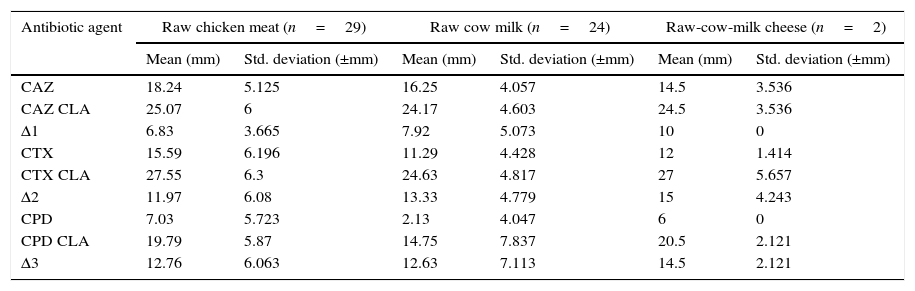
The disc approximation tests were performed by using CPD, CAZ, and CTX±CLA containing discs according to the criteria set by 22. The average zone differences of CAZ±CLA, CTX±CLA and CPD±CLA were found as 6.83±3.66mm, 11.97±6.08mm and 12.76±6.06mm in chicken meat, respectively, and 7.92±5.07mm, 13.33±4.78mm and 12.63±7.11mm in cow milk, respectively, while 10.00±0.00mm, 15.00±4.24mm and 14.50±2.12mm in cow milk cheese, respectively (Table 3).
Zone diameters of CPD, CTX, and CAZ±CLA.
| Antibiotic agent | Raw chicken meat (n=29) | Raw cow milk (n=24) | Raw-cow-milk cheese (n=2) | |||
|---|---|---|---|---|---|---|
| Mean (mm) | Std. deviation (±mm) | Mean (mm) | Std. deviation (±mm) | Mean (mm) | Std. deviation (±mm) | |
| CAZ | 18.24 | 5.125 | 16.25 | 4.057 | 14.5 | 3.536 |
| CAZ CLA | 25.07 | 6 | 24.17 | 4.603 | 24.5 | 3.536 |
| Δ1 | 6.83 | 3.665 | 7.92 | 5.073 | 10 | 0 |
| CTX | 15.59 | 6.196 | 11.29 | 4.428 | 12 | 1.414 |
| CTX CLA | 27.55 | 6.3 | 24.63 | 4.817 | 27 | 5.657 |
| Δ2 | 11.97 | 6.08 | 13.33 | 4.779 | 15 | 4.243 |
| CPD | 7.03 | 5.723 | 2.13 | 4.047 | 6 | 0 |
| CPD CLA | 19.79 | 5.87 | 14.75 | 7.837 | 20.5 | 2.121 |
| Δ3 | 12.76 | 6.063 | 12.63 | 7.113 | 14.5 | 2.121 |
The MIC determination revealed that 62.1% of the isolates from raw chicken meat were resistant to CTX, 55.2% to CAZ, 51.7% to CEP, 20.6% to COX, and 6.9% to CMC, while among the isolates from raw cow milk, 79.2% were resistant to CAZ, 75% to CTX, 66.6% to CEP, 54.2% to COX, 4.2% to ERT, and 4.2% to CMC. Two isolates from raw-cow-milk cheese were 100% resistant to CTX, CAZ, CEP, and CMC.
Among the 55 ESBL producers, five isolates (9.1%) were also positive for AmpC. Based on the species, the AmpC-producing isolates were E. coli (80%) and E. cloacae (20%) from raw chicken meat (n=1), raw cow milk (n=3), and raw-cow-milk cheese (n=1). They were resistant to CAZ (100%), CTX and CEP (80%), CMC (40%), and COX (20%).
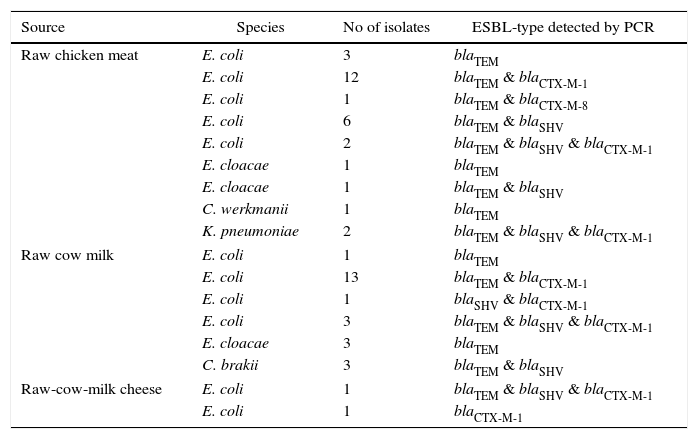
Genotypic resultsIn the genotypic analysis, the PCR products were identified: 53 as blaTEM, 36 as blaCTX-M, and 19 as blaSHV. In the 36 blaCTX-M, 35 were clustered as blaCTX-M-1, and one as blaCTX-M-8. The blaTEM was found in multiple combinations with blaCTX-M-1 & blaCTX-M-8 in 29, with blaSHV in 11, and with blaSHV & blaCTX-M-1 in 7. The blaSHV was detected in a combination with blaCTX-M-1 only in one instance. Of the 44 E. coli isolates, representing the most prevalent ESBL-producing phenotype, a co-existing distribution of bla genes was observed as blaTEM & blaCTX-M-1 in 25, blaSHV & blaTEM in six, blaSHV & blaTEM & blaCTX-M-1 in 6, blaSHV & blaCTX-M-1 in one, blaTEM & blaCTX-M-8 in one, blaTEM alone in four, and blaCTX-M-1 alone in one, respectively (Table 4).
genotypic characteristics of 55 ESBL-producers isolated.
| Source | Species | No of isolates | ESBL-type detected by PCR |
|---|---|---|---|
| Raw chicken meat | E. coli | 3 | blaTEM |
| E. coli | 12 | blaTEM & blaCTX-M-1 | |
| E. coli | 1 | blaTEM & blaCTX-M-8 | |
| E. coli | 6 | blaTEM & blaSHV | |
| E. coli | 2 | blaTEM & blaSHV & blaCTX-M-1 | |
| E. cloacae | 1 | blaTEM | |
| E. cloacae | 1 | blaTEM & blaSHV | |
| C. werkmanii | 1 | blaTEM | |
| K. pneumoniae | 2 | blaTEM & blaSHV & blaCTX-M-1 | |
| Raw cow milk | E. coli | 1 | blaTEM |
| E. coli | 13 | blaTEM & blaCTX-M-1 | |
| E. coli | 1 | blaSHV & blaCTX-M-1 | |
| E. coli | 3 | blaTEM & blaSHV & blaCTX-M-1 | |
| E. cloacae | 3 | blaTEM | |
| C. brakii | 3 | blaTEM & blaSHV | |
| Raw-cow-milk cheese | E. coli | 1 | blaTEM & blaSHV & blaCTX-M-1 |
| E. coli | 1 | blaCTX-M-1 | |
The multiple comparison of the proportions of ESBL producers in their sampling groups was performed using Kruskal–Wallis H-test (H=5.935; P<0.05). The test statistic H was approximated by χ2 distribution (CI 95%; χ2=12.5291; P=0.001). Since H was less than χ2, no statistically significant difference was observed for the association between ESBL proportions and their sampling groups (P<0.05) with respect to the food types of animal origin.
DiscussionIn this study, we screened for ESBL-producing Enterobacteriaceae from raw chicken meat, raw cow milk and raw-cow-milk cheese samples. Subsequently, we characterised genotypically their encoding genes. The results highlighted that 55 isolates were positive for ESBL-producing Enterobacteriaceae; ESBL-producing E. coli (80%) was the most frequent species; a combination of blaTEM & blaCTX-M (52.7%) was the most common pattern; and the major variant of blaCTX-M was defined as blaCTX-M-1 (97.2%), respectively.
The ESBL encoding genetic elements are transferable between the same and different bacterial species.4,25 A study reported that the antibiotic residues in calf urine resulted in amplification of the populations of antibiotic resistant bacteria in the pen soil; and presumably, this increased the probability of transmission of these resistant populations to calves and then the animal-derived food chain.26 This might lead to a risk for infection and colonisation of the human intestinal microbiome with ESBL producing bacteria such as E. coli, E. cloacae, Klebsiella spp., Salmonella spp., Proteus spp. and Citrobacter spp., through various foods of animal origin27–30; when these foods are directly consumed without undergoing any thermal process, or used for raw milk cheese production.31 Due to this threat, the Codex Alimentarius Commission established an intergovernmental task force.32 Following that, in 2014, the Standard Committee of European Doctors (CPME), the Council of European Dentists (CED) and the Federation of Veterinarians of Europe (FVE) issued a common press release for all the authorities to manage the problem of ESBL-producing Enterobacteriaceae (www.fve.org). In our study, a high occurrence of ESBL producers and their encoding genes in the examined foods of animal origin indicate a health risk for the Turkish consumers.
Since the early 2000s, E. coli has become the most concerned member for infection and colonisation of the food animals.33–36 An ESBL-producing E. coli associated mortality was three-times higher than non-ESBL producing E. coli.37 The National Surveillance Network by the Ministry of Health in Turkey (www.uhes.saglik.gov.tr) has reported an increasing prevalence of ESBL-producing E. coli (33.2% in 2008 and 48.83% in 2013) and ESBL-producing K. pneumoniae (40% in 2008 and 49.69% in 2013). However, the role of foods in the dissemination of ESBL-producing bacteria and their encoding genes to humans has not been addressed so far in Turkey. In our study, the frequency of ESBL-producing E. coli was similar to those reported in Portugal (85%)38 and Spain (93.3%),33 while it was higher than those reported in Denmark (1.3%),39 India (3.1%),40 China (12.4%),41 Germany (53.9%),42 Belgium (45%),43 Austria (24%)44 and Taiwan (10.5%).36 Our study also points E. coli being an important foodborne ESBL carrier.
Numerous phenotypic methods have been developed to detect ESBL production by Enterobacteriaceae. However, it is not still clear which tests are the most sensitive.45 In our study, the phenotypic screening and confirmation of ESBL production were performed by the disc approximation tests. All of the used disc combinations showed good sensitivities in accordance with the criteria established by 22. After the disc approximation tests, MIC was determined due to the presence of ESBL encoding genes on the same plasmid.46 The MIC revealed that 55 ESBL producers were resistant to CTX (MIC≥4μg/mL) and CAZ (MIC≥16μg/mL), and partly variable to CPD (MIC≥8μg/mL). Basically, TEM and SHV type beta-lactamases are resistant to CAZ, and variable to CTX while CTX-M type beta-lactamase exhibits resistance to CPD and variable to CAZ because CTX-M type beta-lactamase may escape from screening by CAZ. A survey in Europe reported that up to 33% of ESBLs strains remained undetected, therefore, in this case, genotypic confirmation is further needed.47–50 In our study, the phenotypic results indicated that TEM and CTX-M type beta lactamases were the most commonly encountered ESBLs.
The co-existence of ESBL and AmpC beta-lactamases is a worldwide growing concern.5,51 ESBL and AmpC producing enterobacteria have been clinically reported as 19.5% in Europe, and as 13.9% in Turkey.52,53 In our study, AmpC producers were E. coli and E. cloacae, as reported earlier.52,54,55 Failure to detect these beta lactamases has contributed to their uncontrolled spread and sometimes the consequent therapeutic failures.56 For Enterobacteriaceae, the use of either cefepime (CEP) or cefpirome (CPO)±CLA is recommended since they are un-affected by AmpC.51 In contrast, our results revealed that AmpC producers were 100% resistant to CEP+CLA. This indicates that the techniques to detect AmpC beta-lactamase are still evolving and are not yet optimised.57
The genotypic characterisation by PCR assays revealed that blaTEM was the most frequent gene, followed by blaCTX-M and blaSHV. In Turkey, the clinically reported most frequent beta-lactamase type was CTX-M, followed by TEM and SHV, while another study reported a frequency of SHV higher than TEM.15,58,59 Our genotypic results reporting the most frequent genes as blaTEM & blaCTX-M were also epidemiologically similar to the most commonly encountered ESBL encoding genes in chicken meat from Eastern Europe and Near East,7 Southern America,30,60 Tunisia,61 Northern European countries,33 Japan,62,63 Germany,64 China,35 Austria65 and India,40 followed by the raw milk from Spain,21 England66 and Taiwan36, and by the cheese and dairy products from Portugal,38 England,66 Iran67 and Egypt,68 respectively. However, the foodborne ESBL-producing bacteria and their encoding genes in Turkey have not been addressed till date, and the foodborne beta-lactamases have been studied only by disc approximation testing, excluding genotypic characterisation.69–71 Therefore, our study confirms that the foods of animal origin in Turkey are potential reservoirs of diverse ESBL encoding genes with a co-resistance pattern, which can be disseminated to clinical and community settings.
We detected CTX-M-1 cluster as the most prevalent sub-type of CTX-M type beta lactamases. Chromosomal bla gene, kluC, present in Kluyvera cryocrescens is the ancestor of CTX-M-1 cluster.72 The CTX-M type ESBLs are currently recognised as the part of the most threatening mechanism of antibiotic resistance in clinical and community settings, virtually invading all human and animal compartments as well as the environment all over the world, and are increasingly being predominant form in Enterobacteriaceae worldwide.47,72–74 South America is known as an important source of CTX-M type ESBL originated from clinical settings.75 In Turkey, CTX-M-1 group was found in 86.8% of the clinical E. coli isolates.76 Our results showed that the dissemination of CTX-M-1 cluster was not restricted to the clinical settings, but also involved the foods of animal origin, causing rapid, important and unpredictable changes in the epidemiology of antibiotic resistance.75
In 2007, European Food Safety Authority (EFSA) recommended that bacterial strains containing transferable resistance genes should not be used in animal feeds and fermented and probiotic foods for human use.77,78 However, the issue of antibiotics resistance bacteria in foods has not yet been seriously raised as a health safety indicator. In conclusion, the foods of animal origin sold in Turkey have been found as potential reservoirs for diverse ESBL-producing Enterobacteriaceae and their encoding genes with a co-resistance pattern, thus posing a critical health risk for the Turkish consumers.
DisclaimerThis article is based on the Doctoral thesis entitled “Characterization of Extended Spectrum Beta-Lactamases in Enterobacteriaceae from Foods Using Molecular Method” by İsmail Hakkı Tekiner, 2015, Department of Food Engineering, Institute of Natural and Applied Sciences, Istanbul Aydın University, Istanbul, Turkey.
Conflicts of interestThe authors declare that there is no conflict of interest.