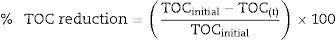Pyrene and benzo[a]pyrene (BaP) are high molecular weight polycyclic aromatic hydrocarbons (PAHs) recalcitrant to microbial attack. Although studies related to the microbial degradation of PAHs have been carried out in the last decades, little is known about degradation of these environmental pollutants by fungi from marine origin. Therefore, this study aimed to select one PAHs degrader among three marine-derived basidiomycete fungi and to study its pyrene detoxification/degradation. Marasmiellus sp. CBMAI 1062 showed higher levels of pyrene and BaP degradation and was subjected to studies related to pyrene degradation optimization using experimental design, acute toxicity, organic carbon removal (TOC), and metabolite evaluation. The experimental design resulted in an efficient pyrene degradation, reducing the experiment time while the PAH concentration applied in the assays was increased. The selected fungus was able to degrade almost 100% of pyrene (0.08mgmL−1) after 48h of incubation under saline condition, without generating toxic compounds and with a TOC reduction of 17%. Intermediate metabolites of pyrene degradation were identified, suggesting that the fungus degraded the compound via the cytochrome P450 system and epoxide hydrolases. These results highlight the relevance of marine-derived fungi in the field of PAH bioremediation, adding value to the blue biotechnology.
Polycyclic aromatic hydrocarbons (PAHs) are organic compounds composed of two or more fused benzene rings. These compounds are formed during the combustion of organic molecules and their subsequent recombination1,2 and are widely distributed in the environment.3,4
PAHs discharged from various activities, such as combustion of fossil fuels, shipping, use and disposal of petroleum products, agricultural burning, coal, and wood-preserving products are persistently hazardous pollutants to the environment.5 Through atmospheric deposition and municipal as well as industrial waste water discharges, PAHs are distributed in the marine environment and finally enriched in the sediment due to their great ability to bind to particulate matter.6
Low molecular weight (LMW) PAHs (composed by 2–3 aromatic rings) are predominant in petrogenic sources and can be introduced into aquatic environments through oil spills, discharge from tanker operations, and municipal and urban runoff. High molecular weight (HMW) PAHs (composed by 4–6 aromatic rings), such as pyrene and BaP, are most important in pyrogenic sources and are released into the environment mainly in the form of exhaust and solid residues.7,8 Both of these compounds are classified by the US Environmental Protection Agency (EPA) as priority pollutants9,10 and have been accumulating in sediments due to their limited water solubility and high affinity for particulate matter.11
Microbial degradation can be considered as an attractive biotechnological alternative for achieving possible mineralization of the pollutant12 and its transformation into less toxic products with greater solubility in water, which can then be degraded by the action of other microorganisms.13,14
Hydrocarbon degradation using fungi from marine origin was reported by Ahearn and Meyers.15 However, their application in this field is still poorly studied. The use of marine-derived fungi for the bioremediation of polluted saline environments2 is facilitated by their tolerance to saline conditions, setting these microorganisms as important microbial resources for biotechnological application in the degradation of PAH polluted environments, such as ocean and marine sediments.16
In the field of environmental pollutants biodegradation studies related to toxicity of metabolites generated during the process is required. For that end, crustaceans and rotifers larvae are applied in ecotoxicological assays due to their ecological relevance and their easy maintenance under laboratory conditions.17–19 Additionally, PAHs biodegradation generally is incomplete, resulting in intermediate compounds. Determinations of PAHs degradation metabolic pathways enable to predict the destiny of such compounds in the environment and elucidate toxicity loss.20,21
Considering that PAHs are pollutants commonly found in the marine environment and that little is known about their degradation by marine microorganism, the present study aimed to screen (among the three marine-derived basidiomycetes) the best fungus able to degrade pyrene and BaP and evaluated its capacity to detoxify and degrade pyrene under saline condition.
Material and methodsMicroorganismsThe marine derived fungal strains Tinctoporellus sp. CBMAI 1061, Marasmiellus sp. CBMAI 1062, and Peniophora sp. CBMAI 1063 were isolated from the Brazilian sponges Dragmacidon reticulatum and Amphimedon viridis collected in the town of São Sebastião, São Paulo State, Brazil.22 All of them were taxonomically identified as previously reported by Bonugli-Santos et al.23 and deposited in the Brazilian Collection of Microorganisms from Environment and Industry – CBMAI (CPQBA, UNICAMP).
Fungal growth and initial screeningThe three isolates were cultivated in 2% (m/v) malt extract agar (MA2) for ten days at 28°C. For initial screening of PAHs degradation (pyrene and BaP), three fungal culture cylinders (0.5cm diameter) from the edge of the colony were transferred to 125mL Erlenmeyer flasks containing 50mL of 2% malt extract broth (MB2) and cultured for 72h at 28°C and 120rpm (initial biomass). Pyrene and BaP were dissolved in dimethylsulfoxide and were added separately at the concentration of 0.04 and 0.02mgmL−1, respectively. The concentration of PAHs applied in this work were chosen based in previous study reported by Passarini.9 The flasks were incubated for 7, 14 and 21 days under the same conditions. No saline conditions were applied for the initial screening.
Quantitative analyses of PAHs degradationPAHs degradation were quantified using gas chromatography–mass spectrometry (GC–MS) analyses, as previously described by Passarini et al.9 The extraction of the PAHs and their degradation products in fermentative medium was accomplished using an Ultra-Turrax system. Ethyl acetate was used as solvent to rupture the fungal cellular walls and remove adsorbed compounds. To each Erlenmeyer flask containing the fungus in fermentative medium, 50mL of ethyl acetate was added, and the fungus was subjected to cellular disintegration at 14,000rpm for 1min. The material was then filtered, transferred to a 250mL separating funnel and shaken vigorously for 1min. The organic phase was filtered by anhydrous sodium sulfate saturated with ethyl acetate directly into a 250mL, round-bottom flask; the aqueous phase was re-extracted with an additional 50mL of ethyl acetate. The organic phases were combined and evaporated under vacuum at 40–45°C until the volume was reduced to approximately 2mL. The contents of the flask were analytically transferred into a 10mL volumetric flask and diluted to the mark with ethyl acetate. An aliquot of 1mL of this solution was then diluted in 10mL of ethyl acetate containing 1mL of the internal standard solution prior to GC–MS analysis.
For the GC–MS analyses, an Agilent (Palo Alto, CA) 6890N GC that was equipped with an Agilent 7683B autosampler coupled to an Agilent 5975 mass-selective detector was used. Data acquisition and analyses were performed using the standard software supplied by the manufacturer. The compounds were separated on a fused-silica capillary column (HP-5MS, 30m×0.25mm i.e., 0.25-μm film thickness) (J&W Scientific, Cologne, Germany). The GC temperature program was as follows: 180°C, 5°Cmin−1 up to 310°C, held for 10min. The temperatures for the injection port and detector were set at 290°C and 300°C, respectively. The splitless injection mode was used with the split outlet opened after 3min, and helium, with a flow rate of 1.0mLmin−1, was used as the carrier gas. The retention times and characteristic mass fragments were recorded, and the chosen diagnostic mass fragments were monitored in the selected ion monitoring (SIM) mode. The characteristic ions that were used for quantification were as follows: benzo[a]pyrene (m/z 252, 126, 250), pyrene (m/z 202, 101, 200) and dibutyl phthalate (m/z 149, 205, 223) as an internal standard. For quantification, the peak area ratios of the analytes to the internal standard were calculated as a function of the compound concentration.
Metabolites identificationThe GC-MS technique was applied for metabolites detection formed during pyrene degradation. The basidiomycete Marasmiellus sp. was incubated in 50mL of liquid medium (MB2) for 5 and 15 days in the presence of pyrene (0.04mgmL−1) at 28°C and 120rpm. The culture medium containing the grown mycelium was extracted in Ultra-Turrax homogenizer using 60mL of dichloromethane as the extractor agent. After two extractions, organic phases were pooled and dried in vacuum using a rotary evaporator in a water bath at 30°C. For each culture time, five replicas were grouped in order to obtain a higher concentration of the formed products. The final residue was dissolved in 2mL of ethyl acetate and 1μL was injected into the GC-MS system. Sample components were separated on a HP-5MS capillary column (30m×0.25mm×0.25μm) following a temperature program: from 60°C to 280°C, up 5°C per minute, kept at 280°C for 26min, and from 280°C to 310°C, up 5°C per minute and held at 310°C for 5min. The injector temperature was 250°C (splitless injection) and the interface temperature was 300°C. Samples were analyzed in Full Scan mode in the 40–600m/z range. Peaks were identified by comparing the mass spectrum and the data available in the library software NIST MS SEARCH version 2.0 and independently through fragmentation pattern interpretation. Samples were also analyzed in the Selected Ion Monitoring mode (SIM). Metabolites were identified by one identification ion and at least one additional confirmation ion per compound, as mass spectra described in literature. Control experiments, without PAH, were also analyzed.
Experimental design and optimization of pyrene degradationIn order to investigate the culture conditions effect on pyrene degradation process by Marasmiellus sp., and to obtain the necessary conditions for efficient degradation, a series of experimental designs were established.24 Initially, the Plackett-Burman design (Matrix with 12 assays) (PB12) was performed for the analyses of the following independent variables evaluation: temperature, pH, salinity and initial concentrations of malt extract, glucose, peptone, yeast extract and PAH. The coded values are presented in Table 2. Salinity was adjusted with artificial seawater that was prepared according to Kester et al.25 All of the different variables were prepared on two levels, designated as −1 (lower) and +1 (upper) (Table 2). Three central point assays were added to the matrix in order to determine the standard error. In a total of 15 trials, the first design allowed to evaluate the effect of 8 variables on PAH degradation after 7 days of incubation. Initial biomass was obtained at 120rpm without the presence of pyrene, then a dimethylsulfoxide solution containing the pollutant was added in separately after 72h of growth. The degradation quantification was performed as previously described. Variables effects on the PAH biodegradation responses were evaluated using the program STATISTICA 7.0.
Based on the results from PB12, a fractional factorial design 24−1 with three central points was purposed to evaluate the effect of four variables on pyrene biodegradation (Table 3): glucose, malt extract, peptone and PAHs after 4 days of incubation. Other growing conditions were 28°C, pH 8, 35ppm salinity and the absence of yeast extract. The variables glucose and peptone were selected to carry out a Central Composite Design 22 (CCD) containing 4 axial points and three repetitions at the central point in order to improve pyrene degradation rates (concentration set at 0.08mgmL−1) after 2 days of incubation (Table 4). The malt extract concentration was set to 25gL−1 and other growth conditions were maintained as described in fractional factorial design 24−1.
After culture conditions optimization for pyrene degradation the metabolites formed were identified as described above. Additionally, acute toxicity and the total organic carbon reduction (TOC) were analyzed as described below.
Acute toxicityArtemia sp. (brine shrimp) dry eggs were purchased from a fish store. Artemia sp. larvae were obtained after hatching from dry eggs in artificial seawater with constant light. Recently hatched (48h) larvae were used for the tests, which were performed in triplicate, using test tubes containing 10% (v/v) NaCl 30% and 4.5mL of samples, totalizing 5mL of test solution. After preparation of the tubes, 30 larvae were added and were exposed to the samples for 24h under light. After that period aspects as Artemia sp. motility and survival were observed. The mortality rate was calculated to determine toxicity and statistics tests (meta-analysis – multiple proportions – p-value <0.01) were run using the software BioStat 5.0.
TOC analysisThe total organic carbon reduction (TOC) analysis was performed with a total organic carbon analyzer, TOC-L Shimadzu, as previously described by Bonugli-Santos et al.26 TOC measurement is based on the combustion of the total carbon (TC) by catalytic oxidation at high temperature. The standard deviation is less than 2% in each sample analyzed in triplicate. The % TOC reduction was calculated using Eq. (1):
where TOCinitial and TOC(t) represent the initial TOC value and the TOC value at time ‘t’ (h), respectively. Samples containing optimized conditions for pyrene degradation in 48h were analyzed. Abiotic controls (without microorganism) were included during the experiment.ResultsPAHs degradation by marine-derived fungiThe three marine-derived basidiomycete fungi were able to degrade pyrene (Fig. 1A) and BaP (Fig. 1B) at different rates. Marasmiellus sp. was able to degrade more than 90% of the initial amount of both pollutants (0.02 and 0.04mgmL−1 BaP and pyrene, respectively) after 7 days of incubation. After 21 days, Tinctoporellus sp. reached almost 50% of degradation, and Peniophora sp. was able to degrade 30% of both PAHs.
Marasmiellus sp. reached high levels of pyrene and BaP degradation after 7 days of incubation and was selected and submitted to additional PAHs degradation studies that were carried out after 1, 3, 5, and 7 days of incubation (Fig. 2). After 24h of incubation, 79.4% of the BaP were degraded, and at the end of the 7th day 97.2% of BaP degradation was achieved. Pyrene was degraded at a slower rate, reaching 18.5% of initial concentration degradation after 24h of incubation. However, after 7 days of incubation 92.8% of pyrene was degraded. Since the results from pyrene degradation were more consistent indicating a degradation curve (Fig. 2), this environmental pollutant was selected and subjected to the studies related to the optimization of degradation, including the analyses of metabolites, toxicity, and total organic carbon reduction (TOC).
Pyrene degradation and detoxification by Marasmiellus sp. CBMAI 1062Analyses through the SIM mode and Full Scan enabled the detection and identification of four metabolites after pyrene degradation by the fungus Marasmiellus sp. (Table 1). Metabolite I presented a molecular ion (M+) m/z 218 and fragment ion m/z 189, resulting in the loss of a –CHO group. It was the most abundant metabolite identified and its standard mass spectrum is characteristic of the hydroxypyrene compound. Metabolites II and III presented molecular ion (M+) m/z 236 and base peak m/z 218. This profile represents H2O unit loss (M+−18). Fragment ion m/z 189, m/z 176 and m/z 94 were also detected. The pattern of the mass spectrum obtained suggests that metabolites II and III are dihydrodiol. The two identified peaks probably correspond to different ring cleavage position. The metabolite IV presented a molecular ion (M+) m/z 234 and fragment ions m/z 205 (M+−COH) and m/z 176 (M+−2COH). The mass spectrum pattern obtained suggests that the metabolite IV is dihydroxypyrene.27 The hydroxyl groups’ position was not determined. None of these metabolites were detected in the experimental control without PAH.
The fungus Marasmiellus sp. was submitted to experimental design studies in order to evaluate the influence of different variables in pyrene degradation and to improve the degradation process. In this context, the influence of 8 independent factors on pyrene degradation by Marasmiellus sp. was investigated using PB12 and the results are shown in Table 2. Pyrene biodegradation ranges from 45.9% to 99.2%. The pH and temperature showed positive effects on pyrene degradation, while the PAH concentration demonstrated a negative effect (Fig. 3). Salinity did not interfere significantly in the pyrene degradation (Fig. 3). However, higher degradation values were observed in saline and alkaline conditions (assays 5 and 7, Table 2).
Plackett-Burman (matrix 12) (PB12) experimental design applied to the evaluation of the effects of 8 factors in pyrene degradation by Marasmiellus sp. CBMAI 1062 after 7 days of incubation.
| Assay | Temperature (°C) | pH | Salinity (ppm) | Glucose (g/50mL) | Malt extract (g/50mL) | Peptone (g/50mL) | Yeast extract (g/50mL) | Pyrene (mgmL−1) | Pyrene degradation (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | +1 (28) | −1 (7) | +1 (35) | −1 (0.25) | −1 (0.25) | −1 (0) | +1 (0.1) | +1 (0.08) | 45.9 |
| 2 | +1 (28) | +1 (9) | −1 (21) | +1 (0.75) | −1 (0.25) | −1 (0) | −1 (0) | +1 (0.08) | 48.9 |
| 3 | −1 (22) | +1 (9) | +1 (35) | −1 (0.25) | +1 (0.75) | −1 (0) | −1 (0) | −1 (0.04) | 91.0 |
| 4 | +1 (28) | −1 (7) | +1 (35) | +1 (0.75) | −1 (0.25) | +1 (0.1) | −1 (0) | −1 (0.04) | 86.2 |
| 5 | +1 (28) | +1 (9) | −1 (21) | +1 (0.75) | +1 (0.75) | −1 (0) | +1 (0.1) | −1 (0.04) | 96.7 |
| 6 | +1 (28) | +1 (9) | +1 (35) | −1 (0.25) | +1 (0.75) | +1 (0.1) | −1 (0) | +1 (0.08) | 82.4 |
| 7 | −1 (22) | +1 (9) | +1 (35) | +1 (0.75) | −1 (0.25) | +1 (0.1) | +1 (0.1) | −1 (0.04) | 93.7 |
| 8 | −1 (22) | −1 (7) | +1 (35) | +1 (0.75) | +1 (0.75) | −1 (0) | +1 (0.1) | +1 (0.08) | 49.4 |
| 9 | −1 (22) | −1 (7) | −1 (21) | +1 (0.75) | +1 (0.75) | +1 (0.1) | −1 (0) | +1 (0.08) | 62.7 |
| 10 | +1 (28) | −1 (7) | −1 (21) | −1 (0.25) | +1 (0.75) | +1 (0.1) | +1 (0.1) | −1 (0.04) | 99.2 |
| 11 | −1 (22) | +1 (9) | −1 (21) | −1 (0.25) | −1 (0.25) | +1 (0.1) | +1 (0.1) | +1 (0.08) | 69.2 |
| 12 | −1 (22) | −1 (7) | −1 (21) | −1 (0.25) | −1 (0.25) | −1 (0) | −1 (0) | −1 (0.04) | 52.8 |
| 13 (C) | 0 (25) | 0 (8) | 0 (28) | 0 (0.5) | 0 (0.5) | 0 (0.05) | 0 (0.05) | 0 (0.06) | 66.6 |
| 14 (C) | 0 (25) | 0 (8) | 0 (28) | 0 (0.5) | 0 (0.5) | 0 (0.05) | 0 (0.05) | 0 (0.06) | 67.3 |
| 15 (C) | 0 (25) | 0 (8) | 0 (28) | 0 (0.5) | 0 (0.5) | 0 (0.05) | 0 (0.05) | 0 (0.06) | 69.0 |
In parenthesis, real values.
Considering the results obtained in the PB12 experiment some variables were chosen for the next stage, a fractional factorial design 24−1: glucose, malt extract, peptone and PAH. Since higher amounts of pyrene were degraded at high rates, the incubation time was reduced from seven to four days. High level of pyrene degradation (98.6%) was also obtained after four days of incubation (assay 8, Table 3), with the three nutrients set at their upper level.
Fractional factorial design 24−1 applied to the evaluation of the effects of 4 factors in pyrene degradation by Marasmiellus sp. CBMAI 1062 after 4 days of incubation.
| Assay | Glucose (g/50mL) | Malt extract (g/50mL) | Peptone (g/50mL) | Pyrene (mgmL−1) | Pyrene degrad. (%) |
|---|---|---|---|---|---|
| 1 | −1 (0) | −1 (0.75) | −1 (0.1) | −1 (0.04) | 94.2 |
| 2 | +1 (0.26) | −1 (0.75) | −1 (0.1) | +1 (0.08) | 92.2 |
| 3 | −1 (0) | +1 (1.25) | −1 (0.1) | +1 (0.08) | 97.8 |
| 4 | +1 (0.26) | +1 (1.25) | −1 (0.1) | −1 (0.04) | 98.0 |
| 5 | −1 (0) | −1 (0.75) | +1 (0.2) | +1 (0.08) | 97.2 |
| 6 | +1 (0.26) | −1 (0.75) | +1 (0.2) | −1 (0.04) | 97.7 |
| 7 | −1 (0) | +1 (1.25) | +1 (0.2) | −1 (0.04) | 98.5 |
| 8 | +1 (0.26) | +1 (1.25) | +1 (0.2) | +1 (0.08) | 98.6 |
| 9 (C) | 0 (0.13) | 0 (1.0) | 0 (0.15) | 0 (0.06) | 95.3 |
| 10 (C) | 0 (0.13) | 0 (1.0) | 0 (0.15) | 0 (0.06) | 96.2 |
| 11 (C) | 0 (0.13) | 0 (1.0) | 0 (0.15) | 0 (0.06) | 97.9 |
In parenthesis, real values.
Since the malt extract had a positive effect on pyrene degradation in the fractional factorial design 24−1 it was set at 25gL−1 (Fig. 4). The PAH concentration was set at 0.08mgmL−1, and incubation time was reduced to two days. Central Composite Design 22 (CCD) was used to evaluate the effects of two factors, glucose and peptone. High rate of degradation (98.2%) was obtained using this strategy (assay 10, Table 4). No significant differences were observed among the assays (listed in Table 4). Thus, it was not possible to propose a mathematical model and to get the response surface.
Central composite design (22) applied to the evaluation of the effects of two factors in pyrene degradation by Marasmiellus sp. CBMAI 1062 after 48h of incubation.
| Assay | Glucose (g/50mL) | Peptone (g/50mL) | Pyrene degradation (%) |
|---|---|---|---|
| 1 | −1 (0.0175) | −1 (0.165) | 92.5 |
| 2 | +1 (0.1025) | −1 (0.165) | 95.1 |
| 3 | −1 (0.0175) | +1 (0.235) | 98.0 |
| 4 | +1 (0.1025) | +1 (0.235) | 97.5 |
| 5 | −1.41 (0) | 0 (0.2) | 96.9 |
| 6 | +1.41 (0.12) | 0 (0.2) | 98.0 |
| 7 | 0 (0.06) | −1.41 (0.15) | 97.9 |
| 8 | 0 (0.06) | +1.41 (0.25) | 97.8 |
| 9 | 0 (0.06) | 0 (0.2) | 97.1 |
| 10 | 0 (0.06) | 0 (0.2) | 98.2 |
| 11 | 0 (0.06) | 0 (0.2) | 98.0 |
In parenthesis, real values.
Assays of the central points, which had the highest pyrene degradation rate were analyzed by GC-MS for the metabolites identification. The same four metabolites previously identified when the fungus was cultured under the non optimized condition (malt extract – non saline condition) after 5 and 15 days of incubation were formed when the fungus were cultured under the optimized condition (malt extract, glucose and peptone – saline condition) after 48h of incubation.
Acute toxicity using Artemia sp. was carried out after the growth of Marasmiellus sp. under pyrene optimized degradation conditions. In the medium containing pyrene (abiotic control) the percentage of survivors was 67.2%, and low motility for the living individuals was observed. In the presence of Marasmiellus sp. and pyrene (bioassay) 95% of survivors with active motility were detected. Statistical tests (meta-analysis – multiple proportions) revealed significant difference between both conditions considering p-value <0.01. Results indicate that there was a loss of toxicity after pyrene biodegradation by Marasmiellus sp.
Studies related to the organic carbon consumption (TOC analysis) were also performed using pyrene optimized degradation conditions. Marasmiellus sp. was able to reduce 17% of TOC after 48h of incubation, indicating that the fungus used the PAHs as carbon sources.
DiscussionData derived from the initial screening revealed the potential of the three marine-derived basidiomycete fungi for PAHs degradation. However, results contradicted data from literature,28 where initial degradation rates were higher for the compound with lower molecular weight (less ring numbers). The toxicity of BaP could result in an induction of BaP degradation by Marasmiellus sp., as a surviving strategy. Additionally, it is worthwhile to mention that the concentration of BaP (0.02mgmL−1) was lower than the pyrene concentration (0.04mgmL−1).
In a previous study reported by our research group9 the best result of PAHs degradation was achieved by Aspergillus sclerotiorum CBMAI 849 that was able to degrade 99.7% of pyrene (0.07mgmL−1) and 76.6% of BaP (0.03mgmL−1) after 8 and 16 days of incubation, respectively. Hadibarata and Kristanti29 also evaluated the pyrene biodegradation by the basidiomycete fungus Armillaria sp. F022 isolated from a tropical rain forest in Indonesia. About 63% of the initial amount of pyrene (0.005mgmL−1) was biodegraded after 30 days of incubation. In another study the maximum pyrene (0.01mgmL−1) degradation rate (99%) was achieved by Pleurotus pulmonarius F043 after 30 days of incubation.5
Considering the results obtained in the screening experiments the fungus Marasmiellus sp. was selected and cultured for 5 and 15 days aiming to detect and identify the metabolites formed during pyrene degradation. The presence of metabolites I, II, III and IV suggest that pyrene was degraded by the cytochrome P450 system and epoxide hydrolases activity. Although the hydroxyl group position was not determined, the 1-hydroxypyrene has been described as a metabolite of fungal pyrene degradation.30,31 The metabolites formed by pyrene biotransformation found in this study were reported as pyrene degradation products by several fungi, including representatives of the genus Marasmiellus.30,32 Lange et al.32 reported that the isolated Marasmiellus ramealis JK 314 was able to degrade 76.5% of pyrene producing 1-hydroxypyrene, 1-pyrenylsulfate, 1,6-hydroxypyrene, 1,6-pyrenequinone and trans-43-dihydro-4,5-dihydroxypyrene metabolites after 14 days of incubation.
It is worth emphasizing that the initial biotransformation of a compound could facilitate the attack by other organisms.33 In addition, the metabolites formed initially as phenols and dihydrodiol can be detoxified by conjugation to other molecules. The conjugates produced by fungal strains include sulfates, glycosides, glucuronides and xilosides.34 These conjugates are generally more soluble in water than the parent compound and can be excreted from the body.
Results from statistical experimental designs showed that efficient pyrene degradation was reached by Marasmiellus sp. in a shorter period of time (48h), higher concentration (0.08mgmL−1), and under saline conditions. Statistical experimental designs can be considered as an efficient approach to determine the best culture conditions in biotechnological processes,35,36 saving cost and time due to the reduction in the total number of experiments.37 Considering that marine-derived fungi are adapted to the ocean conditions, the great performance of the fungus during pyrene degradation under saline condition was expected. However, several studies showed that biodegradation is negatively affected by the increasing in salt concentration.38–40
After pyrene degradation by Marasmiellus sp. there was a loss of toxicity, highlighting the biotechnological potential of this fungus to degrade and detoxify this environmental pollutant. Similar result was reported by Munusamy et al.41 where representatives of the white rot fungus Pycnoporus sanguineus were able to degrade PAH with the production of non toxic compounds based on toxicity analysis using Artemia sp.
Marasmiellus sp. also showed capability to use pyrene as carbon source. Considering the short period of time (48h) applied for pyrene degradation by the fungus, result from TOC reduction can be considered very satisfactory. A higher TOC reduction could be obtained after a longer incubation time. According to Russo et al.42 mushroom compost submitted to combined ozonation and aerobic BaP biodegradation presented from 33% to 72% of TOC removals after 30 days of incubation (soil samples).
ConclusionsResults from the present study revealed the potential of the marine-derived fungi for environmental pollutants degradation. In particular, the fungus Marasmiellus sp. CBMAI 1062 showed great ability to degrade PAHs and can be considered a new microbial resource for bioremediation. Experimental design was successfully applied to optimize the PAH degradation by Marasmiellus sp., resulting in a very high rate (almost 100%) of pyrene degradation after only 48h of incubation. Intermediate metabolites generated suggested that the fungus used the cytochrome P450 system and epoxide hydrolases and highlight the importance of this organism in the early stages of pyrene degradation. Pyrene toxicity was around 28% reduced and total organic carbon was 17% consumed after the degradation process under optimized condition, showing that the fungus was able to detoxify and use the pollutant as carbon source. Additionally, since this fungus was isolated from the marine environment and was able to degrade PAHs under saline conditions, biological advantages can be provided for its use in saline environments and/or processes.
Conflicts of interestThe authors declare no conflicts of interest.
Gabriela A. L. Vieira was supported by a PhD scholarship from the São Paulo Research Foundation – FAPESP (2012/12622-3) and Mariana J. Magrini was supported by a Master's Degree scholarship from the Coordination for the Improvement of Higher Education Personnel – CAPES. The authors thank FAPESP for its financial support (Grants 2010/50190-2, 2013/19486-0 and 2016/07957-7). Lara D. Sette thanks the National Council for Scientific and Technological Development (CNPq) for the Productivity Fellowship (304103/2013-6).