Pasteurella multocida causes atrophic rhinitis in swine and fowl cholera in birds, and is a secondary agent in respiratory syndromes. Pathogenesis and virulence factors involved are still poorly understood. The aim of this study was to detect 22 virulence-associated genes by PCR, including capsular serogroups A, B and D genes and to evaluate the antimicrobial susceptibility of P. multocida strains from poultry and swine. ompH, oma87, plpB, psl, exbD-tonB, fur, hgbA, nanB, sodA, sodC, ptfA were detected in more than 90% of the strains of both hosts. 91% and 92% of avian and swine strains, respectively, were classified in serogroup A. toxA and hsf-1 showed a significant association to serogroup D; pmHAS and pfhA to serogroup A. Gentamicin and amoxicillin were the most effective drugs with susceptibility higher than 97%; however, 76.79% of poultry strains and 85% of swine strains were resistant to sulphonamides. Furthermore, 19.64% and 36.58% of avian and swine strains, respectively, were multi-resistant. Virulence genes studied were not specific to a host and may be the result of horizontal transmission throughout evolution. High multidrug resistance demonstrates the need for responsible use of antimicrobials in animals intended for human consumption, in addition to antimicrobial susceptibility testing to P. multocida.
Pasteurella multocida is a normal inhabitant of the respiratory tract of healthy animals1; however, it is an enigmatic pathogen known for being associated with a diversity of respiratory syndromes that can affect a range of host species.2 Moreover, it is either the primary causative agent or presents a major role in the evolution of certain diseases of economic impact, such as atrophic rhinitis in pigs and fowl cholera (FC) in avian species.2
The variability of the clinical signs in cases of pasteurellosis is possibly explained by the alteration of the commensal relationship between host and bacteria, as well as by the presence of virulence factors that differ several variants of one species of this pathogenic microorganism.1,3 Some molecular studies have been developed to identify genes related to virulence in P. multocida, but little is known regarding the virulence gene content between P. multocida strains in different animal hosts.4–6 The capsule is one of the major virulence factors identified in P. multocida,7 and the genes required for the synthesis and transport of these capsular types are encoded within a single region of the genome.8 All strains can be classified into five different capsular types or serogroups (A, B, D, E and F) according to the presence of capsular antigens.7
Although the strains are generally susceptible to the majority of antimicrobials,9 the increased incidence of multidrug-resistant pathogenic bacteria has been widely reported in the last several decades.6 Compared with human specific pathogens, even less data are available on resistance patterns in pathogens exclusively found in food animals.10 However, the knowledge and the control of antibiotic treatment in P. multocida are important, because the antimicrobial use may select resistant strains, increasing their prevalence.11 Moreover, the use of antibiotics in the control of pasteurellosis, as well as the development of serotype-specific vaccines, requires surveys in each geographical area.12 But studies related to the performance of antimicrobial susceptibility patterns in P. multocida strains of avian origin are rare in Brazil.13 Our objective was to determine whether strains of P. multocida isolated from cases of FC and from pigs with respiratory problems in southern Brazil vary in their repertoire of 22 virulence genes, and to assess the susceptibility of the strains to eight antimicrobial agents.
Materials and methodsStrains of P. multocidaNinety-six strains of P. multocida previously isolated of clinical cases in southern Brazil were selected. After the diagnosis was confirmed, two colonies of each strain plated on blood agar were selected and stored in 1mL of defibrinated sheep blood at a temperature of −80°C. The reactivation and preliminary tests to confirm the presence of pure samples were conducted according to Glisson et al.14 Of the strains selected, 58% (56/96) were isolated from the liver or heart of chickens/turkeys and 42% (40/96) from the lung of pigs that were being raised for human consumption.
Detection of virulence-associated genesAn aliquot of BHI after an overnight incubation (1mL) was selected for DNA extraction using the commercial kit NucleoSpin®Tissue (MachereyNagel®, Düren, Germany). Initially, a PCR protocol for species-specific amplification of the kmt gene was performed, as described by Townsend et al.15 Fifteen genes associated with virulence (ompH, oma87, sodA, sodC, hgbA, hgbB, exBD-tonB, nanB, nanH, ptfA, pfhA, toxA), including the genes for capsule molecular typing of serogroup A (hyaD-hyaC), B (bcbD) and D (dcbF), were surveyed according to the multiplex PCR protocols standardized by Furian et al.16,17 An additional seven genes (hsf-1, pmHAS, fur, psl, ompA, plpB, tadD) were surveyed according to protocols described by Ewers et al.4 and Tang et al.6 with modifications. Reference strains of P. multocida (ATCC 15742, ATCC 12945 and ATCC 12946) were selected as positive controls. Members of the Pasteurellaceae family (Riemerella anatipestifer ATCC 11845, Mannheimia haemolytica ATCC 29694 and Pasteurella gallinarum ATCC 13360) were used as negative controls.
The amplification reactions were performed in a thermocycler (Swift MaxPro Thermal Cycler – ESCO Technologies®, Hatboro, Pennsylvania, USA). Electrophoresis of the amplified products was carried out in a 1% or 1.5% agarose gel (Invitrogen Ultrapure™ Agarose®, Carlsbad, California, USA) stained with ethidium bromide, after which the amplified products were photo documented (AlphaDigDoc Pro – Alpha Innotech®, San Leandro, CA, USA). All protocols were repeated three times in parallel with the relevant positive and negative controls.
Antimicrobial susceptibility testingAll of the strains were tested for their antimicrobial susceptibility by the disk diffusion method according to the performance standards M31–A3 of the Clinical and Laboratory Standards Institute (CLSI).18 Each sample was tested against 8 antimicrobial agents (Oxoid®): erythromycin (15μg), gentamicin (10μg), amoxicillin (10μg), sulphaquinoxaline (300μg), tetracycline (30μg), sulfamethoxazole-trimethoprim (1.25–23.75μg), ceftiofur (30μg) and enrofloxacin (5μg). Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 25923 were selected as quality control strains. The interpretation of results was based on the breakpoints provided by the CLSI guidelines.19,20
Statistical analysisDescriptive statistics were used to calculate the absolute and relative frequencies of the virulence genes and antimicrobial susceptibility profiles. Chi-square (χ2) and Fisher's exact test were used to evaluate the given pair of genes and to determine the association between the genes and the host species of isolation. Comparisons of gene proportions within groups of the same function were made by McNemar's test. Statistical testing was performed with SPSS software (Statistical Package for Social Sciences), and p values of <0.05 were considered statistically significant.
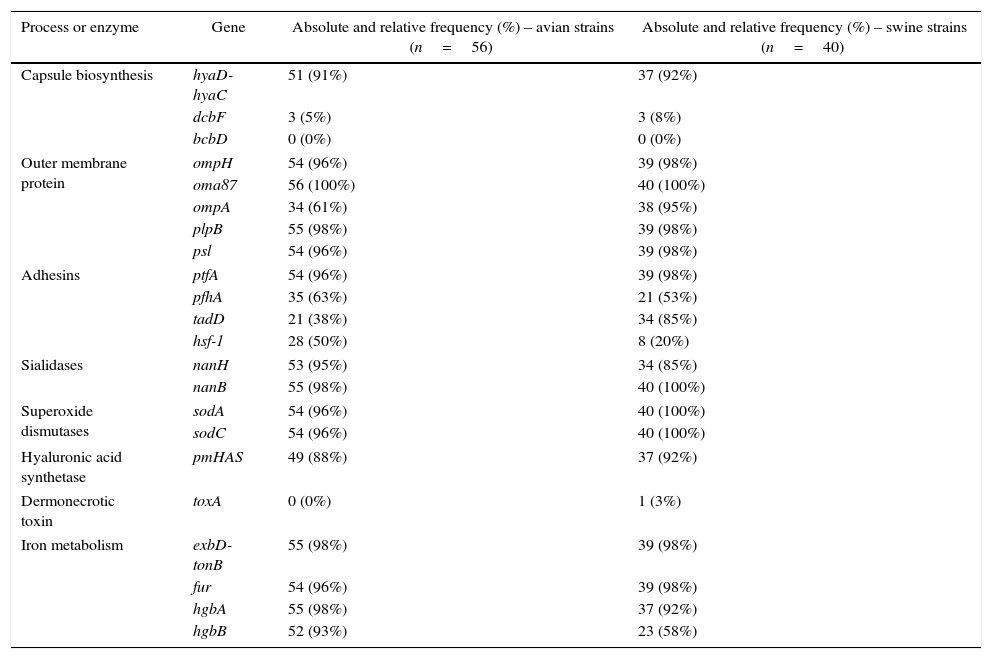
ResultsDistribution of virulence genesThe absolute and relative frequencies of the virulence-associated genes are presented in Table 1. The majority of the virulence-associated genes are regularly distributed among P. multocida strains. The genes associated to outer membrane proteins (ompH, oma87, plpB, psl), adhesion (ptfA), iron mebatolism (exbD-tonB, fur, hgbA), sialidases (nanB) and dismutases (sodA, sodC) were detected in more than 90% of the strains of both hosts.
The distribution of 22 virulence associated genes of Pasteurella multocida detected by PCR according to host species.
| Process or enzyme | Gene | Absolute and relative frequency (%) – avian strains (n=56) | Absolute and relative frequency (%) – swine strains (n=40) |
|---|---|---|---|
| Capsule biosynthesis | hyaD-hyaC | 51 (91%) | 37 (92%) |
| dcbF | 3 (5%) | 3 (8%) | |
| bcbD | 0 (0%) | 0 (0%) | |
| Outer membrane protein | ompH | 54 (96%) | 39 (98%) |
| oma87 | 56 (100%) | 40 (100%) | |
| ompA | 34 (61%) | 38 (95%) | |
| plpB | 55 (98%) | 39 (98%) | |
| psl | 54 (96%) | 39 (98%) | |
| Adhesins | ptfA | 54 (96%) | 39 (98%) |
| pfhA | 35 (63%) | 21 (53%) | |
| tadD | 21 (38%) | 34 (85%) | |
| hsf-1 | 28 (50%) | 8 (20%) | |
| Sialidases | nanH | 53 (95%) | 34 (85%) |
| nanB | 55 (98%) | 40 (100%) | |
| Superoxide dismutases | sodA | 54 (96%) | 40 (100%) |
| sodC | 54 (96%) | 40 (100%) | |
| Hyaluronic acid synthetase | pmHAS | 49 (88%) | 37 (92%) |
| Dermonecrotic toxin | toxA | 0 (0%) | 1 (3%) |
| Iron metabolism | exbD-tonB | 55 (98%) | 39 (98%) |
| fur | 54 (96%) | 39 (98%) | |
| hgbA | 55 (98%) | 37 (92%) | |
| hgbB | 52 (93%) | 23 (58%) | |
The genes hyaD-hyaC, dcbF and bcbD are associated with capsular biosynthesis of P. multocida specific to serogroups A, D and B, respectively. Over 90% of the strains belonged to serogroup A in both host species, and there was a strong significant difference compared to dcbF (p<0.001). Two strains of avian origin were not identified in any of the three capsular types. More than 90% of the strains were positive for genes encoding outer membrane proteins (ompH, oma87, psl, plpB) in both host species. However, ompA was detected in 61% of avian strains with a significantly lower occurrence (p<0.001) than that observed for the other genes in the same functional group. Similarly, hgbB showed a lower percentage (58%) than other genes involved in iron metabolism among the swine strains (p<0.001). No significant difference was observed between exbD-tonB, fur and hgbA (p>0.05). The genes encoding sialidases (nanH, nanB), dismutases (sodA, sodC) and hyaluronan synthase (pmHAS) also showed frequencies close to or higher than 90% and differences within the groups were not significant (p>0.05).
A higher variation of frequencies was observed among the adhesins. The gene ptfA encoding a subunit of type IV fimbriae was significantly higher in both host species (p<0.001) compared to pfhA and hsf-1, which encode a filamentous hemagglutinin and an autotransport adhesin, respectively. Additionally, the frequency of pfhA exhibited a significant difference (p<0.05) compared to tadD in both hosts. On the other hand, a significant difference between tadD and hsf-1 frequencies (p<0.001) was only observed among strains isolated from pigs.
The association between virulence genes and serogroupsThe association between virulence genes and serogroups is presented in Table 2. Some virulence genes exhibited distinctive associations with serogroup A and serogroup D. The genes toxA and hsf-1 are positively associated with serogroup D, and the genes pmHAS and pfhA with serogroup A. On the other hand, a negative association of pmHAS, pfhA and tadD with serogroup D was observed, as well as of hsf-1 with serogroup A.
The absolute and relative frequencies (%) of virulence associated genes detected by PCR according to serogroups A and D of Pasteurella multocida.
| Gene | Serogroup A (gene hyaD-hyaC) (n=88) | Serogroup D (gene dcbF) (n=6) |
|---|---|---|
| ompH | 85 (97%) | 6 (100%) |
| ompA | 65 (74%) | 6 (100%) |
| plpB | 86 (98%) | 6 (100%) |
| psl | 86 (98%) | 5 (83%) |
| exbD-tonB | 86 (98%) | 6 (100%) |
| fur | 85 (97%) | 6 (100%) |
| hgbA | 84 (95%) | 6 (100%) |
| hgbB | 67 (76%) | 6 (100%) |
| nanH | 79 (90%) | 6 (100%) |
| nanB | 87 (99%) | 6 (100%) |
| sodA | 86 (98%) | 6 (100%) |
| sodC | 86 (98%) | 6 (100%) |
| pmHAS | 86 (98%)** | 0 (0%)** |
| toxA | 0 (0%) | 1 (17%)* |
| ptfA | 85 (97%) | 6 (100%) |
| pfhA | 56 (64%)* | 0 (0%)* |
| tadD | 53 (60%) | 0 (0%)* |
| hsf-1 | 28 (32%)** | 6 (100%)* |
oma87: constant in both serogroups.
The combination of genes among P. multocida regardless of host species of isolation is shown in Table 3. In addition to the high frequencies exhibited by the majority of selected genes, there was a significant association in the combination of virulence genes in 45 situations. The combinations between plpB and exbD-tonB, tadD and hgbB, and ompA and hsf-1 indicate a strong significant association (p<0.001), as shown in Table 3. Each of the genes, except tadD, presented a significant association (p<0.001) for both host species and in this case cannot be selected as species-specific markers.
The percentage of virulence-associated genes in pairs among 96 strains of Pasteurella multocida analyzed.
| ompH | toxA | ptfA | nanH | exbD-tonB | sodA | pfhA | hgbA | sodC | nanB | hgbB | oma87 | hsf-1 | fur | pmHAS | psI | ompA | plpB | tadA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ompH | 100 | ||||||||||||||||||
| toxA | 1 | 100 | |||||||||||||||||
| ptfA | 97 | 1 | 100 | ||||||||||||||||
| nanH | 93* | 100 | 90 | 100 | |||||||||||||||
| exbD-tonB | 100* | 100 | 98 | 100* | 100 | ||||||||||||||
| sodA | 98 | 100 | 98 | 99 | 98 | 100 | |||||||||||||
| pfhA | 58 | 0 | 57 | 60 | 59 | 59 | 100 | ||||||||||||
| hgbA | 96 | 100 | 96 | 99* | 96 | 96 | 100* | 100 | |||||||||||
| sodC | 99 | 1 | 98 | 99 | 99* | 98 | 100 | 98 | 100 | ||||||||||
| nanB | 100* | 100 | 99 | 100 | 100* | 99 | 100 | 99 | 100* | 100 | |||||||||
| hgbB | 78 | 100 | 78 | 80 | 78 | 78 | 77 | 82* | 78 | 78 | 100 | ||||||||
| oma87 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |||||||
| hsf-1 | 36* | 100 | 38 | 33* | 36 | 37 | 38 | 35* | 36 | 37 | 42 | 38 | 100 | ||||||
| fur | 99* | 100 | 97 | 99* | 99* | 97 | 96 | 97 | 98 | 98* | 96 | 97 | 92* | 100 | |||||
| pmHAS | 90 | 0 | 90 | 89 | 89 | 89 | 98* | 90 | 89 | 90 | 87 | 90 | 78* | 90 | 100 | ||||
| psl | 99* | 100 | 97 | 99* | 99* | 97 | 98 | 97 | 98 | 98* | 96 | 97 | 92* | 99* | 98 | 100 | |||
| ompA | 77* | 100 | 76 | 76* | 44 | 75 | 63* | 74 | 77 | 76 | 68* | 75 | 39** | 77* | 73 | 76 | 100 | ||
| plpB | 100* | 100 | 98 | 100* | 100** | 98 | 98 | 98 | 99 | 99* | 97 | 98 | 94 | 100* | 98 | 100* | 100 | 100 | |
| tadD | 60 | 0 | 58 | 57* | 59 | 56 | 48* | 55 | 59 | 58 | 45** | 57 | 28* | 59* | 62* | 59 | 74* | 59 | 100 |
The association of hyaD-hyaC and dcbF with other genes is listed in Table 2.
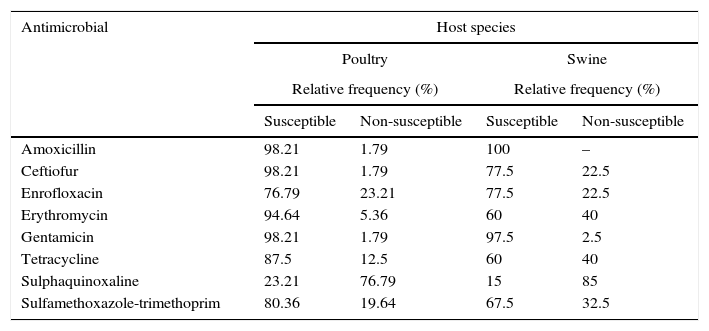
The antimicrobial susceptibility patterns of P. multocida strains are described in Table 4. No drug was effective in inhibiting the growth of 100% of the poultry strains, but strains of this group showed a higher susceptibility to 80% of the antimicrobials tested, with the exception of enrofloxacin and sulphaquinoxaline. Only amoxicillin and gentamicin inhibited the growth of more than 80% of porcine strains. For the purpose of this study, multidrug-resistance (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories.21 36.58% of the porcine strains were multi-resistant, while 19.64% of the poultry strains were resistant to three or more drugs in different categories. The highest percentages of resistance observed, regardless of the host origin, were related to sulphaquinoxaline.
The antimicrobial susceptibility patterns of Pasteurella multocida strains by disk diffusion test according to host species.
| Antimicrobial | Host species | |||
|---|---|---|---|---|
| Poultry | Swine | |||
| Relative frequency (%) | Relative frequency (%) | |||
| Susceptible | Non-susceptible | Susceptible | Non-susceptible | |
| Amoxicillin | 98.21 | 1.79 | 100 | – |
| Ceftiofur | 98.21 | 1.79 | 77.5 | 22.5 |
| Enrofloxacin | 76.79 | 23.21 | 77.5 | 22.5 |
| Erythromycin | 94.64 | 5.36 | 60 | 40 |
| Gentamicin | 98.21 | 1.79 | 97.5 | 2.5 |
| Tetracycline | 87.5 | 12.5 | 60 | 40 |
| Sulphaquinoxaline | 23.21 | 76.79 | 15 | 85 |
| Sulfamethoxazole-trimethoprim | 80.36 | 19.64 | 67.5 | 32.5 |
The serogroup classification of P. multocida according the capsular typing is used for studies on the pathogenesis and epidemiology of the agent. The biological basis of this phenomenon is unknown, but it suggests that the capsule is related to the pathogenesis of the individual diseases and to host predilection of particular serogroups.22 It is known that in addition to the specific relation to certain diseases and species, there is a dominant geographical distribution of the capsular types in some cases. However, these relations have shown variations in recent years.23 Serogroup classification is rare in Brazil, especially with strains of P. multocida isolated from poultry. Type A, identified in 91% of the avian strains, is the major serogroup found in this host, and similar results were observed in other studies.24,25 Only 5% of the avian strains were classified in capsular types D and none in type B, which is also present in birds, as well as type F. The occurrence of these serogroups is considered rare.26 Likewise, capsular type A was also prevalent among strains from swine (92%), which demonstrates a variable prevalence of the serogroups in cases of respiratory disease according to the geographic region.6,27
Although the majority of virulence genes presented a similar distribution between serogroups, some were significantly associated with a specific capsular type, such as pfhA with serogroup A. Tang et al.6 reported the presence of pfhA in 25% of strains from serogroup A, and only in 3.1% of the strains from type D. Similar results were obtained by Ewers et al.4 that analyzed 289 strains isolated from various host species. The positive association of pfhA and capA is not likely due to a physical linkage, as the genes are separated by approximately 839kb in the genome of P. multocida strain Pm70. Hypothetically, a capA positive clone may have acquired pfhA by horizontal gene transfer and achieved an advantage to survive within the host.5 The toxA gene, encoding a dermonecrotic toxin (PMT – P. multocida toxin), presented a significant association to serogroup D (p<0.05). This toxin induces osteolysis in the turbinate bones and plays an important role in the pathogenesis of progressive atrophic rhinitis.27 Although toxA is associated with serogroup D,28 other studies have detected it in strains of serogroup A isolated from different hosts.29 Tang et al.6 also observed the positive association of hsf-1 with serogroup D and the negative association of pmHAS with the same serogroup. On the other hand, pmHAS was significantly associated with serogroup A. The hyaluronan synthase PmHAS is a dual-action synthase that transfers alternating units of d-glucuronic acid and N-acetyl-d-glucosamine in the formation of hyaluronic acid, the major type A capsular material.30
The high frequency of virulence genes analyzed was also observed in other studies with strains from both hosts.4–6 A similar distribution of certain genes, regardless of host species or P. multocida serotype, may suggest the selection of factors that present cross-protection as candidates for vaccine development. Some of these antigens potentially serve as vaccine candidates.31,32 As an example, porins are generally conserved among species and have high immunogenicity.33 However, studies with experimental infection frequently show variable results in animal protection.33,34
The ability of pathogens to obtain iron is a key feature during the infection process.35 The frequency of hgbB in avian strains was higher than the occurrence of 85% observed for Ewers et al.,4 the only study that investigated its presence in this host. On the other hand, the gene was detected in only 58% of the strains from swine. Bethe et al.5 detected hgbB in less frequency in diseased than in healthy animals, a result also found by Shayegh et al.,29 who analyzed strains from ovine. In both studies, there is not thought to be a relationship between the presence of hgbB and pasteurellosis.
The majority of commensal and pathogenic bacteria that interact with eukaryotic hosts express adhesive molecules on their surfaces to promote interaction with host cell receptors.36 With the exception of ptfA that encodes a fimbrial subunit type IV,37 the other adhesin-related genes presented a frequency inferior to 65%. Studies show a variation in the frequency of adhesins and differ in interpretation regarding the relationship between the adhesin and the disease. Ewers et al.4 observed a variation between 7 and 100% of pfhA according to the host species and related its presence to the occurrence of pasteurellosis in cattle. Similarly, Bethe et al.5 connected pfhA with the occurrence of respiratory disease in swine. However, Shayegh et al.29 detected a low frequency (18%) of the gene among ovine strains and did not relate it with pasteurellosis in this species.
Antibiotic therapy is still an effective tool in the treatment of infections caused by P. multocida, as the strains were susceptible to the majority of drugs commercially available. The high susceptibility of strains from both hosts to amoxicillin, gentamicin and ceftiofur was also observed in other studies in different countries,6,9,38,39 as well as in Brazil with strains of porcine origin.40,41 In contrast to other studies that report low activity of aminoglycosides,38 gentamicin was effective in the current study. Likewise, resistance to erythromycin generally observed42 was not found among the strains from poultry.
However, excessive and unjustified use of antimicrobials may accelerate the emergence of resistant strains. Among the conventional drugs in veterinary medicine, tetracyclines are widely employed,11,12 which explains the high percentage of resistance in this study and the literature.11,43 Another example is the high frequency of non-susceptible strains to quinolones and the low efficacy of sulphonamides, chemotherapeutic agents indicated for the treatment of FC,43 also found in several studies.6,42,44
Increasing multidrug resistance is attributable at least in part to the use of antibiotic additives in animal feed and the extensive use of antimicrobial agents in veterinary medicine.6 Similarly, the horizontal transfer of genes by mobile genetic elements from different bacterial genera and species favors the development of multidrug-resistance.45 The presence of P. multocida strains in a polymicrobial environment offers the opportunity to acquire resistance genes that have developed in other bacteria but have been refined under selective pressure.10
ConclusionThis study provided information regarding the distribution of virulence genes in strains of P. multocida isolated from poultry and swine in Brazil. These genes presented a high frequency in the majority of the cases and they were not specific to a host. Similarly, high multidrug resistance evinces the need of restrictive and responsible use of antimicrobials in animals intended for human consumption, in addition to antimicrobial susceptibility testing to P. multocida.
Conflicts of interestThe authors declare no conflicts of interest.