Paraoxonase 1 (PON1) plays a major role in the oxidation of low density lipoprotein and in the prevention of coronary atherogenesis. In this context, coding region polymorphisms of PON1 gene, responsible for the enzyme activity, has become of interest as a marker for atherogenesis.
MethodsA study and follow-up was conducted on 529 patients with an acute coronary event in order to assess the association between the PON1 Q192R (rs662;A/G) polymorphism, the type of acute coronary syndrome, cardiovascular risk factors (arterial hypertension, diabetes mellitus, dyslipidaemia, and smoking), the extent and severity of coronary atherosclerosis, and the medium-term clinical follow-up.
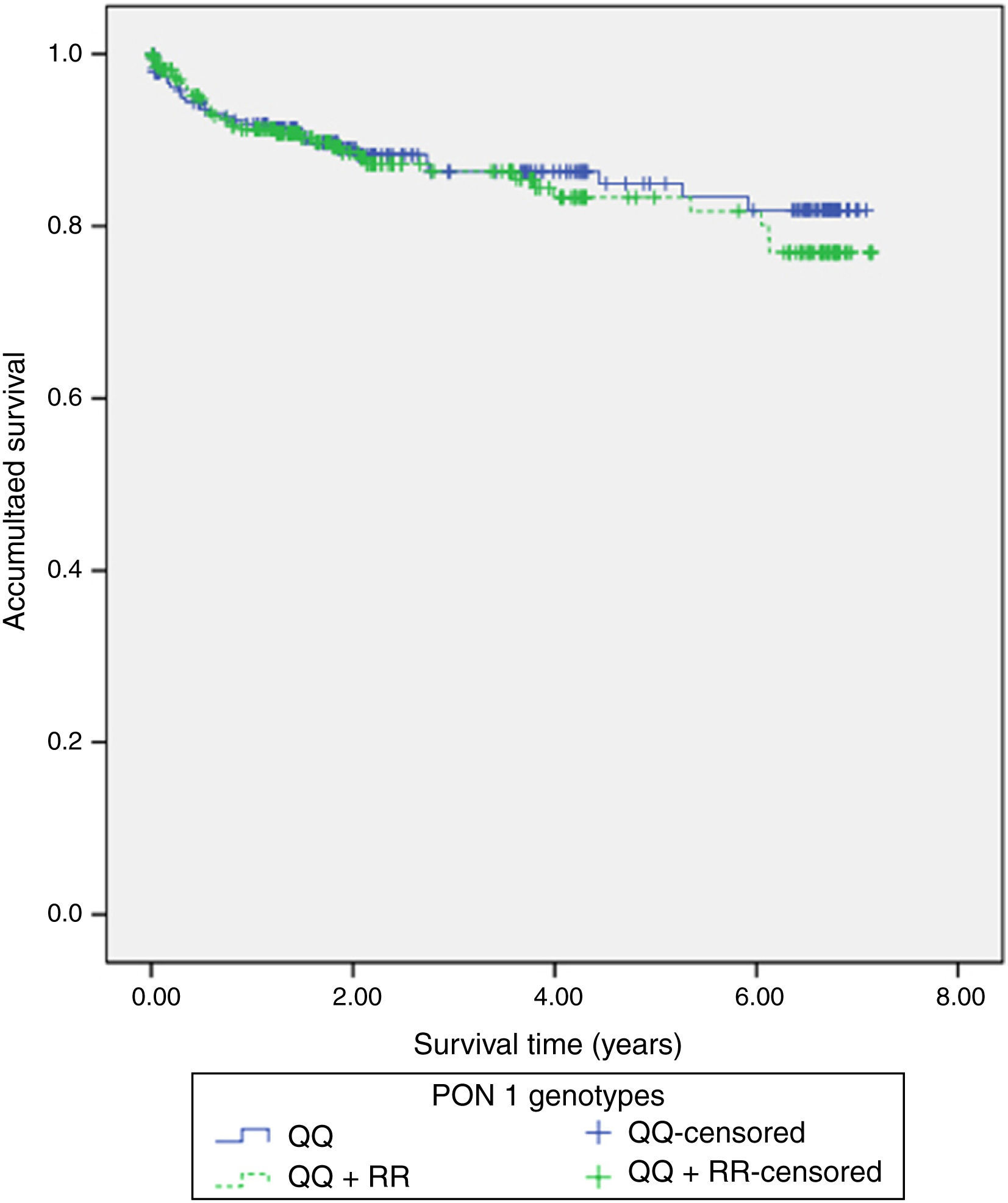
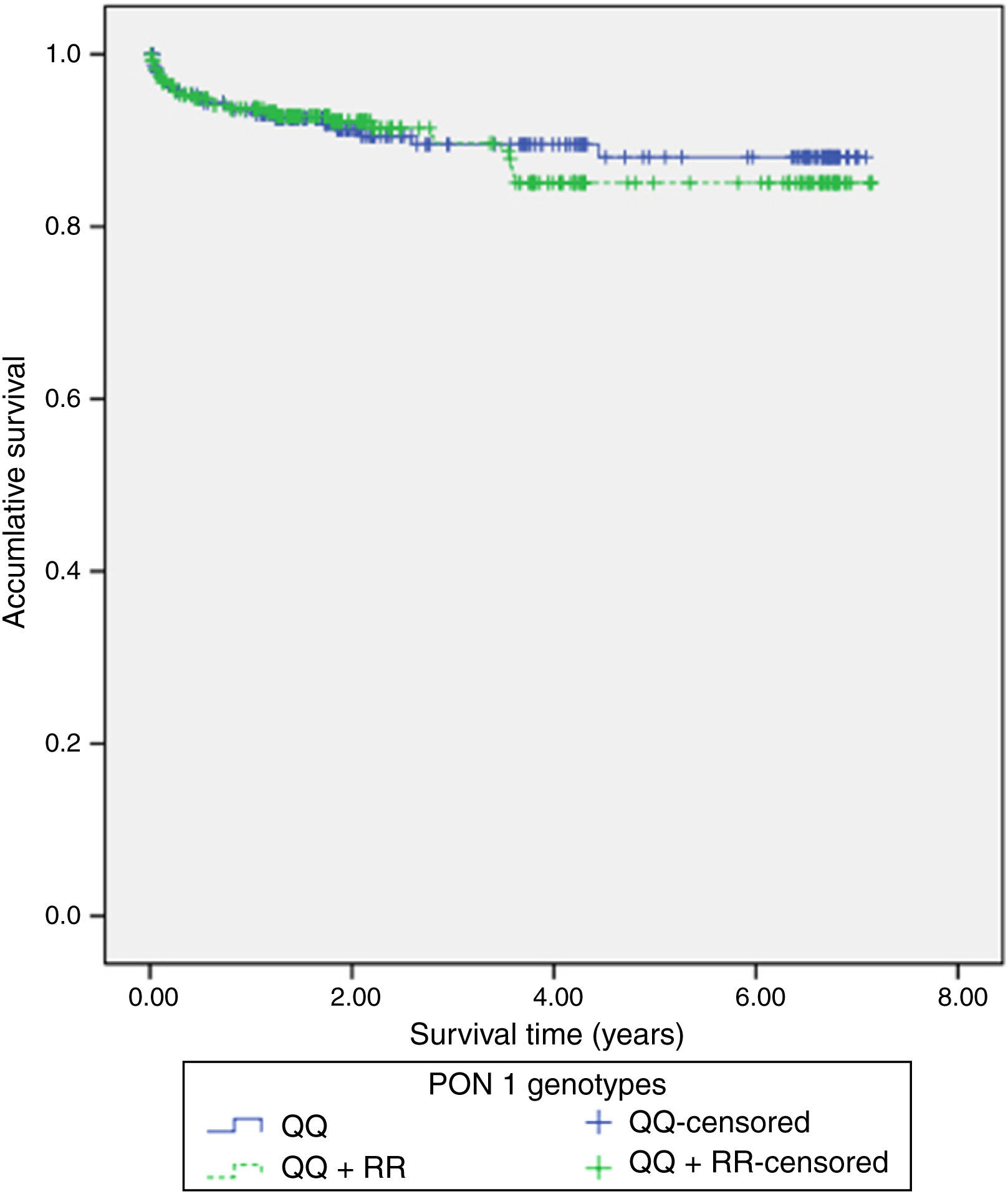
ResultsThe QQ genotype was found in 245 (46.3%) patients, with 218 (41.2%) patients showing the QR genotype, and 66 (14.5%) patients had the RR genotype. No significant differences were found between the QQ and QR/RR genotypes as regards the clinical characteristics, the analytical data, and the angiographic variables. Similarly, Kaplan–Meier survival analysis showed no significant differences in presenting with a new acute coronary event (p=0.598), cardiac mortality (p=0.701), stent thrombosis (p=0.508), or stent re-stenosis (p=0.598) between QQ and QR/RR genotypes during the follow-up period (3.3±2.2 years).
ConclusionsIn patients with an acute coronary syndrome, the PON1 Q192R genotypes did not influence the risk of suffering a new acute coronary event during the medium-term follow-up.
La paraoxonasa 1 (PON1) juega un papel importante en la oxidación de las lipoproteínas de baja densidad y en la prevención de la aterosclerosis coronaria. En este contexto, el gen de la PON1, responsable de la actividad de la enzima, se ha convertido en un marcador de interés del proceso aterogénico.
MétodosSe estudiaron y siguieron a 529 pacientes con síndrome coronario agudo para evaluar la asociación entre el polimorfismo Q192R (rs662; A/G) del PON1 y el tipo de evento coronario, los factores de riesgo cardiovascular (hipertensión arterial, diabetes mellitus, dislipidemia y tabaquismo), la extensión y gravedad de la aterosclerosis coronaria y la incidencia de eventos cardiovasculares a medio plazo.
ResultadosUn total de 245 pacientes (46,3%) presentaron el genotipo QQ, 218 (41,2%) el genotipo QR y 66 (14,5%) el genotipo RR. No se encontraron diferencias significativas entre el genotipo QQ y los genotipos QR/RR en relación con las variables clínicas, analíticas y angiográficas estudiadas. Por su parte, el análisis de Kaplan-Meier de supervivencia no mostró diferencias significativas en relación con la reincidencia de nuevos eventos coronarios (p=0,598), la mortalidad cardíaca (p=0,701), la trombosis del stent (p=0,508) o la reestenosis del stent (p=0,598) entre los genotipos QQ vs. QR/RR durante el tiempo de seguimiento (3,3±2,2 años).
ConclusionesEl genotipo PON1 Q192R no influyó en el riesgo de sufrir un reevento coronario agudo a medio plazo entre los pacientes que presentaron un síndrome coronario agudo.
Serum paraoxonase-1 (PON1) is a high-density lipoprotein (HDL)-associated enzyme, synthesized and secreted by the liver.1 In human serum, most if not all of the paraoxonase activity is associated with HDL. The antiatherogenic properties of serum paraoxonase-1 includes a reduced macrophage oxidative status and a smaller oxidation of low-density lipoprotein (LDL),2 a decreased stimulation cholesterol efflux from macrophages, a declined monocyte chemotaxis/adhesion to endothelial cells and an inhibition of monocyte to macrophage differentiation.3 Therefore a genetic polymorphism that results in a lower enzyme activity may lead to an increased incidence of arteriosclerosis.
This is the case the Q192R (rs662; A/G) polymorphism of the PON1 gene. The R allele (arginine at position 192) displays several-fold higher activity toward paraoxon hydrolysis than the Q allele (glutamine at position 192), which means a lower PON1 activity in the serum.2 In fact, in knockout mice lacking the PON1 gene, atherosclerosis develops more rapidly than in wild-type mice, whereas mice that over express human PON1 are resistant to atherosclerosis.4,5 In such context, it has been suggested that the Q allele, which is more abundant than the R allele, is responsible for the protective effect against atherosclerosis, whereas the R allele has been related to coronary heart disease.
The aim of this study is to assess if the PON1 Q192R polymorphism is associated not only to the type of coronary event and the severity of coronary atherosclerosis d but also to the relapse of a new coronary event.
MethodsSubjectsA retrospective study of 529 consecutive patients suffering an acute coronary event, between January 2009 and September 2010, was carried out. None of them had intercurrent inflammatory disease, fever, known malignancies or liver failure. All patients were Caucasian, at least 18 years old and all of them gave informed consent before being included in the study, which was approved by the clinical research ethics committee of our institution. A review of the patient's past medical history was done in all the patients.
The in-hospital data at admission included: type of acute coronary event (unstable angina, Non-ST-Elevation Myocardial Infarction (NSTEMI) and ST-elevation myocardial infarction (STEMI)), type of stent (bare-metal or drug-eluting type), analytical data at admission and pharmacological treatment at discharge (aspirin, clopidogrel, prasugrel, beta adrenoreceptor antagonists, angiotensin converting enzyme inhibitors, angiotensin receptor blockers, calcium channel blocker, nitrates or statins). Also hospital readmission from any cause (stable angina, unstable angina, NSTEMI, STEMI, arrhythmias or heart failure) and total mortality was determined. The diagnosis of an acute coronary syndrome was done by clinical history, electrocardiogram and serial measurements of troponin I. Myocardial infarction was diagnosed when troponin I serum levels were above 0.16ng/dL. The left ventricular ejection fraction (LVEF) was calculated by echocardiography using Simpson's biplane method.6
Evaluation of cardiovascular risk factorsArterial hypertension was diagnosed when the patient presented with systolic blood pressure above 140mmHg or diastolic blood pressure above 90mmHg, or used medication prescribed for hypertension; diabetes mellitus, if the patient reported to be diabetic (fasting blood glucose levels higher than 126mg/dL) or was treated with oral anti-diabetic agents or insulin; dyslipidemia, if the patient presented with total serum cholesterol levels above 240mg/dL, or was with lipid-lowering therapy, and smoking if the patients was an active smoker at admission or in the last 2 years, or non-smoker if the patient had never smoked nor smoked for the last two years before admission. The family history of coronary heart disease was obtained by interviewing participants whether any first-degree relatives had experienced a fatal or nonfatal myocardial infarction and/or coronary angioplasty/coronary artery bypass surgery. Any cardiovascular event was considered premature if it occurred before the age of 55 in males and 65 in females.
Analytical dataAfter fasting for at least 10h, blood samples were drawn for the measurements of serum glucose, serum creatinine, and total cholesterol, that were determined by spectrophotometric methods using an AU 2700 (Olympus Diagnostic, Hamburg, Germany). Troponin I levels were determined with the VIDAS Troponin I Ultra assay system (bioMerieux, Marcy. L’Etoil, France).
Coronary angiographyCoronary angiography was performed using standard angiographic techniques.
Angiographic scoring was carried out by interventional cardiologists who were blinded to the study protocol. Significant coronary artery disease was defined as the presence of any luminal stenosis >75%. Based on visual angiographic results, patients were divided into 0, 1, 2 or 3 vessels disease depending on the number of coronary vessels with significant stenosis.
Genetic analysisGenomic DNA was extracted from venous blood by using the salting out procedure. The Q192R variant of the PON1 gene (SNP ID: rs662) was detected by using a Restriction Fragment Length Polymorphism (RFLP) assay as previously described.7
Statistical analysisVariables are presented as mean±standard deviation, counts (percentages), or median (interquartile range). Kolmogorov-Smirnov test was used to check for normal distribution of continuous data. Categorical variables were compared using ×2 test. Comparisons among continuous variables were assessed using two-sided unpaired t-student or Mann–Whitney U tests depending on the data distribution. A one-way ANOVA was used to compare the means of more than two independent groups. Calculations for endpoints of interest were based on the comparison of the PON1 allele carriage status among subjects with the QQ genotype and carriers of the polymorphic allele (QR/RR). A dominant model was chosen because the polymorphic */R alleles results in a lower enzyme function activity, which means that one mutant allele exerts a relevant effect on overall enzyme activity. The overall free-survival rates were calculated using Kaplan–Meier method, and the differences determined using the log rank test. All p-values<0.05 were accepted as statistically significant. Statistical analysis was performed using SPSS 17.0 for Windows (SPSS Inc., Chicago, IL, United States).
ResultsThe allele frequency of the rs662 SNP, defining the Q192R variant (529) was as follows: 245 (46.3%) patients were QQ homozygotes, 218 (41.2%) patients were QR heterozygotes and 66 (14.5%) patients were homozygotes for the RR genotype, proportions that were similar to what is reported for this variant in other European populations.7 Therefore all genotypes were well represented in our sample population.
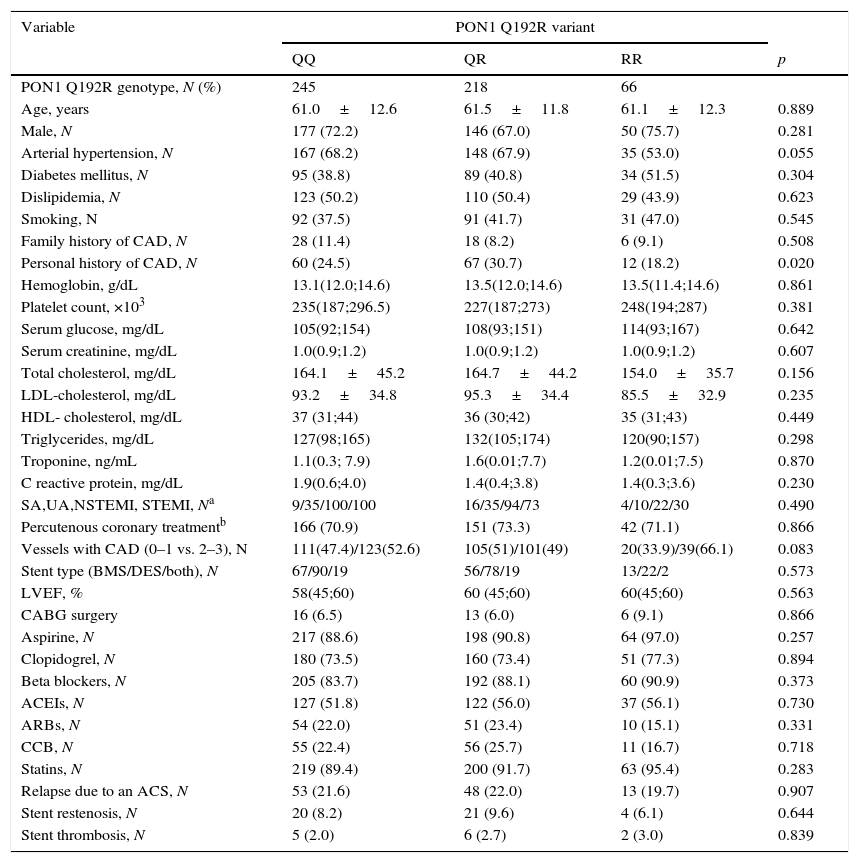
Table 1 shows the demographic, clinical, analytical, angiographic/echocardiographic data and percutaneous/medical treatment at hospital discharge of the different genotypes. No significance was found between them except in the presence of personal history of coronary artery disease. However, when the QQ genotype was compared with the combination of the QR/RR genotypes none of the parameters studied in Table 1 reached statistical significance.
Demographic, clinical, analytical and follow up data in the different PON1 Q192R genotypes.
| Variable | PON1 Q192R variant | |||
|---|---|---|---|---|
| QR | RR | p | ||
| PON1 Q192R genotype, N (%) | 245 | 218 | 66 | |
| Age, years | 61.0±12.6 | 61.5±11.8 | 61.1±12.3 | 0.889 |
| Male, N | 177 (72.2) | 146 (67.0) | 50 (75.7) | 0.281 |
| Arterial hypertension, N | 167 (68.2) | 148 (67.9) | 35 (53.0) | 0.055 |
| Diabetes mellitus, N | 95 (38.8) | 89 (40.8) | 34 (51.5) | 0.304 |
| Dislipidemia, N | 123 (50.2) | 110 (50.4) | 29 (43.9) | 0.623 |
| Smoking, N | 92 (37.5) | 91 (41.7) | 31 (47.0) | 0.545 |
| Family history of CAD, N | 28 (11.4) | 18 (8.2) | 6 (9.1) | 0.508 |
| Personal history of CAD, N | 60 (24.5) | 67 (30.7) | 12 (18.2) | 0.020 |
| Hemoglobin, g/dL | 13.1(12.0;14.6) | 13.5(12.0;14.6) | 13.5(11.4;14.6) | 0.861 |
| Platelet count, ×103 | 235(187;296.5) | 227(187;273) | 248(194;287) | 0.381 |
| Serum glucose, mg/dL | 105(92;154) | 108(93;151) | 114(93;167) | 0.642 |
| Serum creatinine, mg/dL | 1.0(0.9;1.2) | 1.0(0.9;1.2) | 1.0(0.9;1.2) | 0.607 |
| Total cholesterol, mg/dL | 164.1±45.2 | 164.7±44.2 | 154.0±35.7 | 0.156 |
| LDL-cholesterol, mg/dL | 93.2±34.8 | 95.3±34.4 | 85.5±32.9 | 0.235 |
| HDL- cholesterol, mg/dL | 37 (31;44) | 36 (30;42) | 35 (31;43) | 0.449 |
| Triglycerides, mg/dL | 127(98;165) | 132(105;174) | 120(90;157) | 0.298 |
| Troponine, ng/mL | 1.1(0.3; 7.9) | 1.6(0.01;7.7) | 1.2(0.01;7.5) | 0.870 |
| C reactive protein, mg/dL | 1.9(0.6;4.0) | 1.4(0.4;3.8) | 1.4(0.3;3.6) | 0.230 |
| SA,UA,NSTEMI, STEMI, Na | 9/35/100/100 | 16/35/94/73 | 4/10/22/30 | 0.490 |
| Percutenous coronary treatmentb | 166 (70.9) | 151 (73.3) | 42 (71.1) | 0.866 |
| Vessels with CAD (0–1 vs. 2–3), N | 111(47.4)/123(52.6) | 105(51)/101(49) | 20(33.9)/39(66.1) | 0.083 |
| Stent type (BMS/DES/both), N | 67/90/19 | 56/78/19 | 13/22/2 | 0.573 |
| LVEF, % | 58(45;60) | 60 (45;60) | 60(45;60) | 0.563 |
| CABG surgery | 16 (6.5) | 13 (6.0) | 6 (9.1) | 0.866 |
| Aspirine, N | 217 (88.6) | 198 (90.8) | 64 (97.0) | 0.257 |
| Clopidogrel, N | 180 (73.5) | 160 (73.4) | 51 (77.3) | 0.894 |
| Beta blockers, N | 205 (83.7) | 192 (88.1) | 60 (90.9) | 0.373 |
| ACEIs, N | 127 (51.8) | 122 (56.0) | 37 (56.1) | 0.730 |
| ARBs, N | 54 (22.0) | 51 (23.4) | 10 (15.1) | 0.331 |
| CCB, N | 55 (22.4) | 56 (25.7) | 11 (16.7) | 0.718 |
| Statins, N | 219 (89.4) | 200 (91.7) | 63 (95.4) | 0.283 |
| Relapse due to an ACS, N | 53 (21.6) | 48 (22.0) | 13 (19.7) | 0.907 |
| Stent restenosis, N | 20 (8.2) | 21 (9.6) | 4 (6.1) | 0.644 |
| Stent thrombosis, N | 5 (2.0) | 6 (2.7) | 2 (3.0) | 0.839 |
Data are shown as means±standard deviation, medians and interquartiles (25th–75th percentile) or frequencies (%). ACEIs: angiotensin-converting enzyme inhibitors, ACS: acute coronary syndrome, ARBs: Angiotensin II receptor blockers, BMS: bare metal stent, CABG: coronary artery bypass grafting, CAD: coronary artery disease, CCB: calcium channel blockers, DES: drug eluting stent, HDL: High-density lipoprotein, LDL: Low-density lipoprotein, LVEF: left ventricular ejection fraction, N: number of patients, NSTEMI: non ST elevation myocardial infarction, PON1: paraoxonase-1, SA: stable angina, STEMI; ST elevation myocardial infarction, UA: unstable angina.
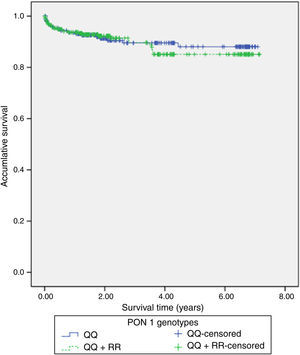
117 (22%) patients had hospital readmissions due to cardiovascular events during the follow-up (3.3±2.2 years): 33 patients had a NSTEMI, 27 heart failure, 23 unstable angina, 16 stable angina, 12 STEMI, 5 cerebrovascular events and 1 patient a n arrhythmic event. Among those 68 patients with a new acute coronary syndrome (of which 65 patients underwent coronary angiography) 13 (20%) patients showed stent reestenosis, 5 (8%) patients had stent thrombosis and 47 (72%) patients developed de novo coronary lesions. Among those 117 patients who needed hospital readmission, 41 (35%) died during the follow up: 14 patients had in-hospital cardiac death, 16 patients had out-of-hospital death and 11 patients had non-cardiovascular mortality. Kaplan–Meier survival analysis showed no significant differences in presenting a new coronary syndrome (p=0.598) (Fig. 1), cardiac mortality (p=0.701) (Fig. 2), stent thrombosis (p=0.508) or stent reestenosis (p=0.598) between QQ versus QR/RR genotypes during the follow up time.
The Q192R (rs662; A/G) polymorphism, which results in the glutamine to arginine substitution at position 192 of the PON1 gene, has been associated with a decreased PON1 activity and an increased atherosclerosis risk in several but not all population studies. Some authors have found a positive relationship between coronary heart disease and the RR genotype8–11 while others have not.12–14 Similarly, the R allele has been associated with an increased risk for cardiovascular disease in some populations at high risk,15 but not in the general population at large.16 In such context, a meta-analysis of 43 studies showed a weak association between the RR genotype and the presence of coronary disease17 although not a single study has found the R allele to be less commonly associated with coronary heart disease18 than the Q allele. These data are consistent with the findings seen in our series where no significant differences were seen in angiographic coronary stenosis between the QQ vs. the QR/RR genotypes.
Although PON1 activity and concentration are determined genetically, various factors, such as diet, lifestyle, and environmental factors, can influence PON1 activity and/or concentration. For example, smoking decreases serum PON1 activity19 while moderate alcohol intake increases it.20 Moreover, activity and concentration of PON1 can vary up to 40-fold in human populations.21 In such context, some authors have evidenced that PON1 enzyme activity is lower in subjects with significant coronary artery disease, and that there is a significant relationship between PON1 activity and coronary atherosclerosis determined by coronary angiography.22
In our series we found no significant differences between the QQ and QR/RR genotypes neither in clinical or analytical variables nor in the extent of coronary artery disease. However, when the three alleles were taken separately, we found a higher incidence of personal history of coronary artery disease among those patients with the QQ genotype which could be also in relation to the most frequent association with systemic arterial hypertension.23
Similarly, we did not find any significant difference, during the follow up time, between the QQ and the QR/RR genotypes and the relapase of a new coronary syndrome or cardiac death. As the RR genotype has a role in coronary artery disease through the oxidation of lipids, which is a key factor for coronary plaque destabilization, medical treatment with antiplatelet drugs and statins added to the high number of patients who were treated percutaneously in both genotypes groups may have influenced the absence of significance. In addition, some authors have recently shown a poor correlation of this polymorphism with the oxidation of lipoprotein lipids24 which may have contributed also to the lack of significance of our results.
We acknowledge the limitations of our study as other factors may limit PON1 activity besides the genotype. Also, lipid peroxides which are substrates for PON1 and which have been shown to be raised in people with coronary heart disease are inhibitors of PON1.25 In addition, proatherogenic diets and a wide interindividual variation in blood of paraoxonase activity may limit its value.26 In fact, both PON1 activity and genotype taken together appear as better predictors of atherosclerotic risk than both taken separately.27
In conclusion, in patients suffering an acute coronary event the PON1 Q192R genotypes did not influence the risk of suffering a new relapse during the medium term follow up. However, these results may have been influenced by the high percentage of patients under dual antiplatelet therapy, beta blockers and statins treatment and the high number of patients who were treated percutaneously in our series.
Conflict of interestThere is no conflict of interest by the authors. None of the authors has received funding. All authors participated in the interpretation of data, drafting the article and final approval of the version to be published.
Ethical disclosureProtection of human and animal subjectsThe authors declare that the procedures followed were in accordance with the regulations of the responsible Clinical Research Ethics Committee and in accordance with those of the World Medical Association and the Helsinki Declaration.
Confidentiality of dataThe authors declare that they have followed the protocols of their work centre on the publication of patient data.
Right to privacy and informed consentThe authors must have obtained the informed consent of the patients and/or subjects mentioned in the article. The author for correspondence must be in possession of this document.