Salmonella is an enteropathogen acquired through contaminated food or water. In Colombia, Salmonella spp. is included in the national surveillance of Acute Diarrhoeal Diseases and typhoid fever initiated in 1997. This report shows the phenotype and genotype results obtained from 2005 to 2011.
MethodsA total of 4010 isolates of Salmonella enterica were analysed by serotyping with Kauffmann–White–LeMinor, antimicrobial resistance patterns, and pulse-field gel electrophoresis (PFGE).
ResultsA total of 93 serovars were identified, of which, Typhimurium, Enteritidis, Typhi, Dublin, Panama, Derby, Braenderup, Saintpaul, and Uganda were prominent. The highest levels of resistance were found for tetracycline and nalidixic acid. Susceptibility was observed in 52.4% (2101/4010) of the isolates. Multi-resistance was recorded in 54.9% of Typhimurium isolates, with 81 different combinations. Using PFGE, 51.9% (2083/4010) isolates were analysed in 34 serovars, and 828 electrophoretic patterns were obtained. From these, 8 patterns were found in at least two Latin-American countries.
ConclusionThe surveillance of Salmonella spp. provides information on the serovar distribution, antimicrobial resistance, and clonal distribution in Colombia, as well as information to treat this disease and control the spread of antimicrobial bacterial resistance.
Salmonella spp. es un enteropatógeno que se transmite a los humanos a través de alimentos o agua contaminada. En 1997, el Grupo de Microbiología del Instituto Nacional de Salud de Colombia inició el programa de vigilancia de enfermedad diarreica aguda y fiebre tifoidea, que incluye Salmonella spp. Este informe presenta los resultados fenotípicos y genotípicos de los aislamientos recuperados de 2005 a 2011 como parte de la vigilancia.
MétodosUn total de 4.010 aislamientos de Salmonella spp. fueron analizados por serotipificación con el esquema Kauffmann-White-LeMinor, patrones de sensibilidad antimicrobiana y de electroforesis en gel de campos pulsados (PFGE).
ResultadosSe identificaron un total de 93 serovares, con 9 predominantes, Typhimurium, Enteritidis, Typhi, Dublin, Panama, Derby, Braenderup, Saintpaul y Uganda. Salmonella spp. presentó altos porcentajes de resistencia a tetraciclina y ácido nalidíxico. El 52,4% (2.101/4.010) de los aislamientos fueron sensibles a todos los antibióticos. La multirresistencia se observó en el 54,9% de los aislamientos de Typhimurium, representada por 81 combinaciones. Por PFGE se analizaron 51,9% aislamientos (2.083/4.010) de 34 serovares, generando 828 patrones electroforéticos XbaI. De estos, 8 se reportaron en al menos 2 países en Latinoamérica.
ConclusiónLa vigilancia de Salmonella spp. permite conocer la distribución de los serovares, su resistencia y la identificación de clones endémicos en Colombia, aportando bases para un tratamiento óptimo en las infecciones generadas por este patógeno y en el diseño de programas para disminuir la dispersión de aislamientos multirresistentes.
Salmonella spp. is an enteropathogen that is passed on to humans through contaminated food or water and is thus classified as a foodborne illness (FBI).1 People infected with Salmonella spp. present symptoms such as diarrhoea, fever and abdominal pain. It primarily affects children under 5 years of age, causing acute diarrhoeal disease (ADD).2 In the United States, 19,056 cases, 4200 hospital admissions and 80 deaths due to FBI were reported in 2013.3 As such, reducing infection has been one of the five priority goals of the US Department of Health and Human Services since 2012. These measures are the result of information generated by laboratory surveillance, which serves as guide to direct efforts for FBI prevention.3,4 In Colombia, the ADD mortality rate among the general population is 1.57 per 100,000 population, with a fatality of 0.04% for 2011. It is more common in children under 10 years of age but the risk of death is higher in patients over 80 years.5
More than 2600 serovars have been described in two species of Salmonella spp.: S. bongori and S. enterica. The latter is subdivided into seven subspecies, and the pathogenic serovars that infect humans belong to this enterica subspecies. Serovars Typhimurium and Enteritidis are recovered worldwide from gastroenteritis symptoms and have a wide range of hosts, while serovars Typhi (for which humans are the only host), Sendai and Paratyphi A, B and C cause typhoid fever.6–8
The US Centers for Disease Control and Prevention estimates that Salmonella spp. causes one million cases of disease, with 19,000 hospital admissions and 380 deaths per year.1
In Colombia, national Salmonella spp. surveillance was established from 1997 as an information network that includes the shipment of isolates from the country's hospitals to 32 Departmental Public Health Laboratories (PHLs) and the Capital District. One of these is the Microbiology Group of the National Institute of Health (NIH), where phenotypic characterisation is performed by means of serotyping and antimicrobial and genotypic sensitivity profiling, determined by pulsed-field gel electrophoresis (PFGE).
This report presents the results obtained in the phenotypic and genotypic surveillance of Salmonella spp. isolates from human clinical samples, from 2005 to 2011, within the ADD and typhoid fever surveillance programme.
Materials and methodsPhenotypic characterisationThe Salmonella spp. isolates sent to the NIH Microbiology Group were confirmed through biochemical testing9 and serotyping, following the Kauffmann–White–Le Minor scheme.10
The antimicrobial susceptibility profiles were determined using the disc diffusion method (Kirby–Bauer), following the recommendations and interpretation criteria of the Clinical and Laboratory Standards Institute (CLSI)11 for tetracycline (TE), chloramphenicol (C), nalidixic acid (NA), amoxicillin/clavulanic acid (AMC), aztreonam (AZT), amikacin (AK) and streptomycin (S); the latter three antibiotics were only evaluated for serovar Typhimurium. The minimum inhibitory concentration (MIC) was determined in the AutoSCAN-4 device (Siemens, Germany) with the NC50 panel for ampicillin (AMP), trimethoprim/sulfamethoxazole (SXT), ciprofloxacin (CIP), cefotaxime (CTX) and ceftazidime (CAZ). The isolates that were sensitive to all of the antibiotics were defined as pansensitive and those with a resistance to three or more families of antibiotics were multidrug-resistant (MDR).
Serovars with a total number of isolates greater than or equal to 60 in the 7 years under study were considered predominant within Colombia.
Genotypic characterisationGenotypic characterisation by PFGE was performed on a percentage of all of the isolates, following the procedures and guidelines established by the PulseNet Latin America and Caribbean Network (PulseNet-LA and Caribbean).12 The isolates had to meet one of the following criteria: being serovar Typhi; being from common and uncommon serovars that present unusual characteristics related to the sample type and resistance profile; and being recovered in outbreaks. Serovars Panama and Saintpaul were processed following the PulseNet network thiourea protocol.
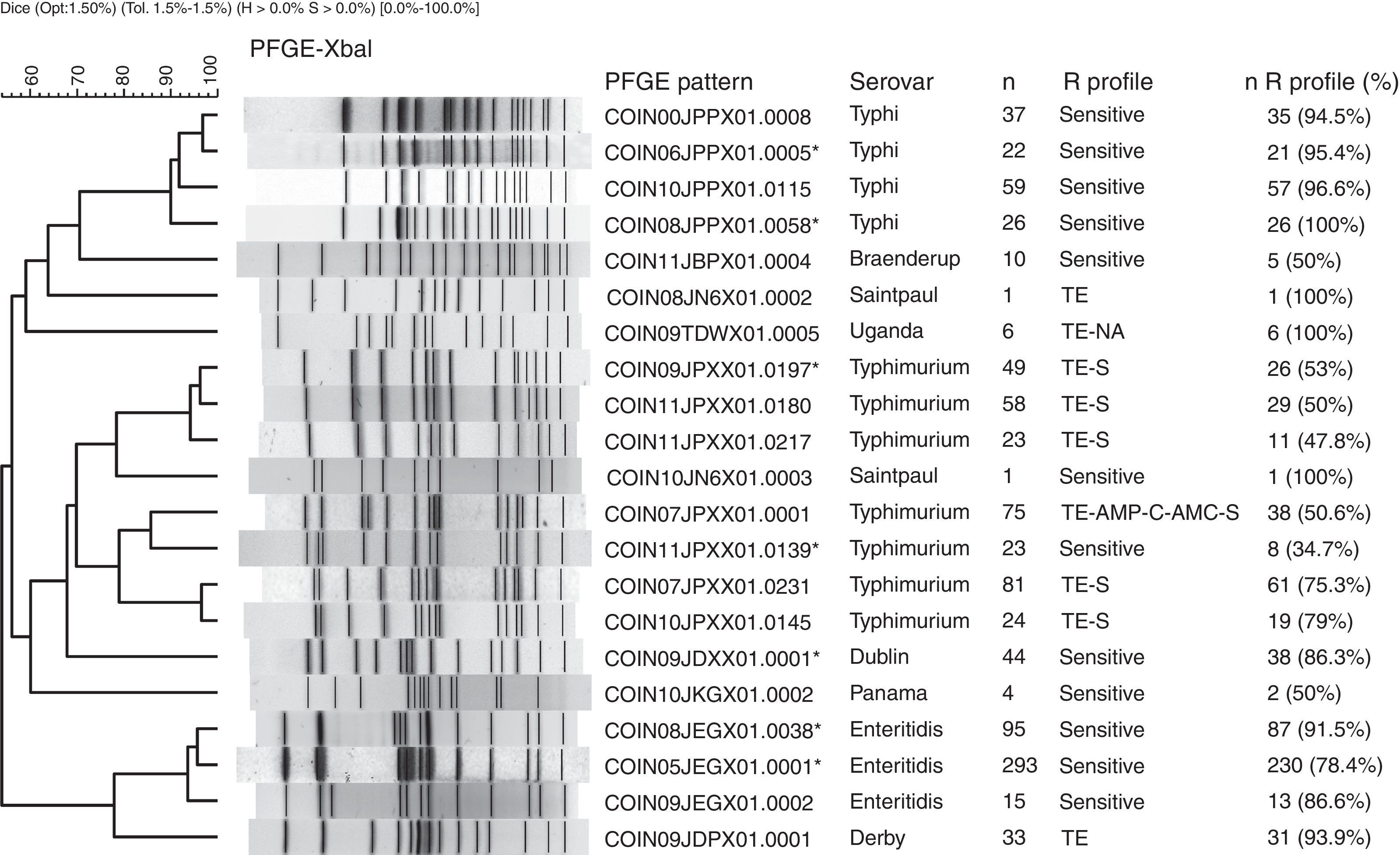
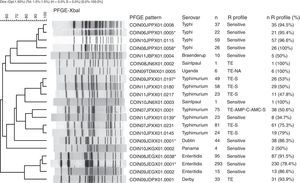
The PFGE pattern obtained for each isolate was compared to the national database, which comprises a representative of each PFGE pattern found in the different serovars. The PFGE patterns were named according to the previously established PulseNet network parameters13 (Fig. 1). The patterns from the national database were then compared to the PulseNet-LA and Caribbean regional database (RDB), where electrophoretic profiles of Salmonella enterica from the 17 countries participating in the Network can be found.14 This comparison allows for the identification of patterns shared with one or more Latin American countries on presenting 100% genetic similarity.
Main PFGE patterns in the 9 predominant Salmonella spp. serovars in Colombia. The dendrogram generated with the UPGMA algorithm is shown, using the Dice coefficient with 1.5% tolerance. The PFGE patterns are named according to the previously established PulseNet network parameters.13
CO: Colombia, IN: National Institute of Health, 2 numbers indicate the year the isolate was recovered, 3 capital letters identify each serovar, 3 alphanumeric characters correspond to the restriction enzyme (for Xbal this is X01) and the final 4 numbers correspond to the consecutively assigned pattern. The patterns marked with an asterisk are those that share a 100% likeness with the RDB patterns. The following column indicates the serovar, followed by the number of isolates with the respective PFGE pattern (n). The R Profile column shows the predominant resistance profile in each PFGE pattern. The n R profile (%) column shows the number of isolates with the predominant resistance profile, and the n R profile/n percentage was calculated.
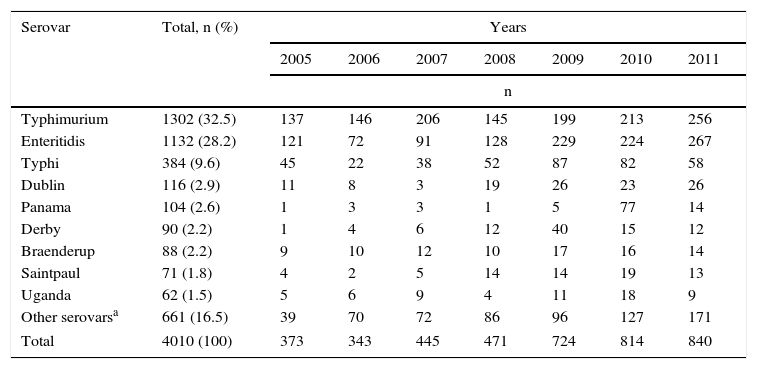
In the 7 years under analysis, 4010 Salmonella spp. isolates were received by the Microbiology Group, from which 92 serovars were identified. Three serovars made up 70.3% of the isolates: Typhimurium with 32.5%, Enteritidis with 28.2% and Typhi with 9.6%. The following six serovars made up 13.2% of the isolates, and the remaining 83 serovars made up 16.5%, with each of them having fewer than five isolates (Table 1).
Most frequently recovered Salmonella spp. serovars in Colombia from 2005 to 2011.
| Serovar | Total, n (%) | Years | ||||||
|---|---|---|---|---|---|---|---|---|
| 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | ||
| n | ||||||||
| Typhimurium | 1302 (32.5) | 137 | 146 | 206 | 145 | 199 | 213 | 256 |
| Enteritidis | 1132 (28.2) | 121 | 72 | 91 | 128 | 229 | 224 | 267 |
| Typhi | 384 (9.6) | 45 | 22 | 38 | 52 | 87 | 82 | 58 |
| Dublin | 116 (2.9) | 11 | 8 | 3 | 19 | 26 | 23 | 26 |
| Panama | 104 (2.6) | 1 | 3 | 3 | 1 | 5 | 77 | 14 |
| Derby | 90 (2.2) | 1 | 4 | 6 | 12 | 40 | 15 | 12 |
| Braenderup | 88 (2.2) | 9 | 10 | 12 | 10 | 17 | 16 | 14 |
| Saintpaul | 71 (1.8) | 4 | 2 | 5 | 14 | 14 | 19 | 13 |
| Uganda | 62 (1.5) | 5 | 6 | 9 | 4 | 11 | 18 | 9 |
| Other serovarsa | 661 (16.5) | 39 | 70 | 72 | 86 | 96 | 127 | 171 |
| Total | 4010 (100) | 373 | 343 | 445 | 471 | 724 | 814 | 840 |
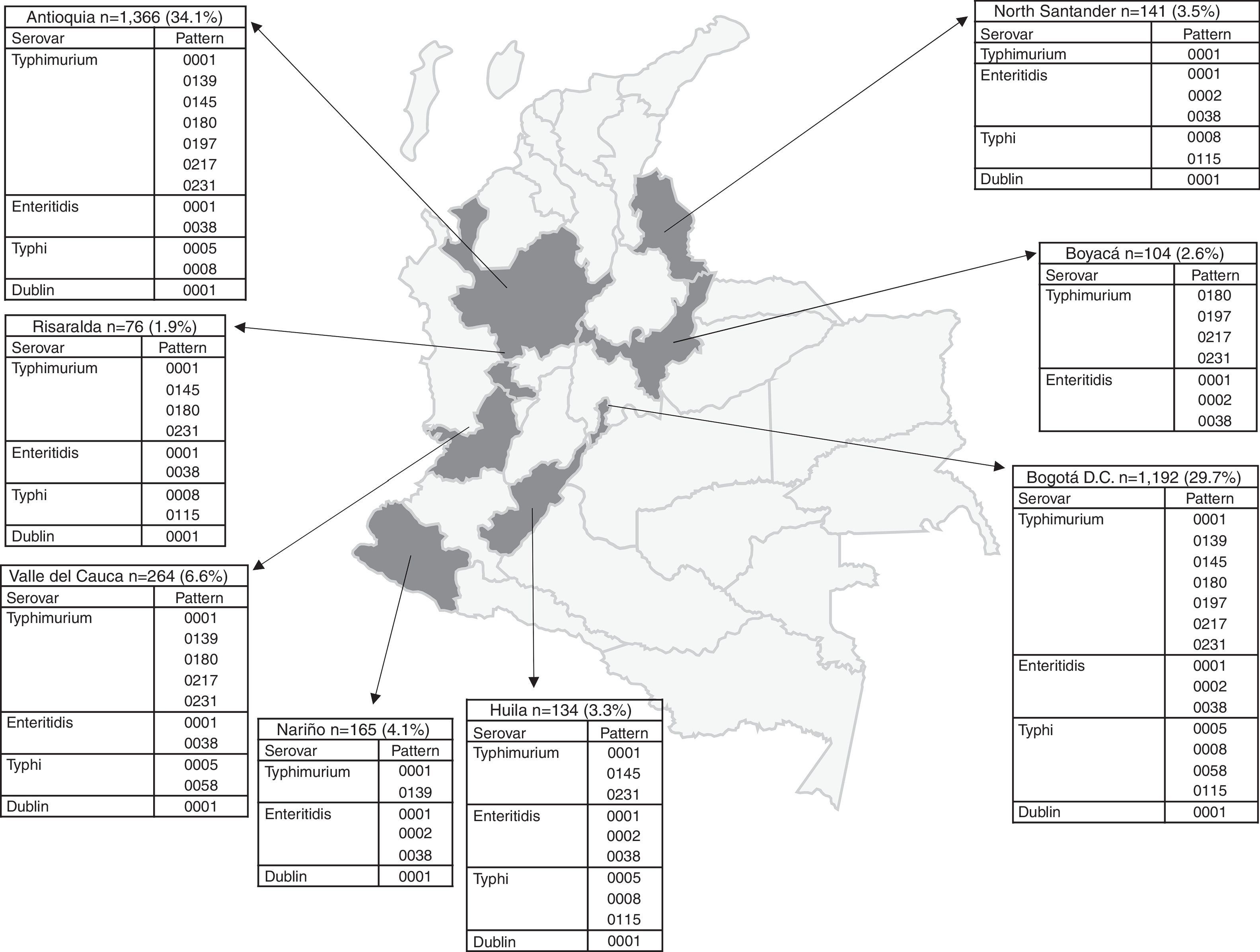
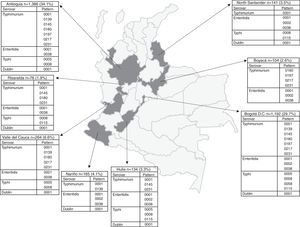
The number of Salmonella spp. isolates received increased year-on-year, from 373 in 2005 to 840 in 2011 (Table 1). The department of Antioquia and the Capital District of Bogotá provided 63.8% of the isolates, with 1366 (34.1%) and 1192 (29.7%), respectively, followed by the departments of Valle, Nariño and Santander, with 264 (6.6%), 165 (4.1%) and 141 (3.5%) isolates, respectively (Fig. 2). Serovar Typhimurium was predominant until 2008, when it was overtaken by Enteritidis. In the years 2008–2010, an increase in the number of serovar Typhi isolates was observed (Table 1).
Departmental distribution of the predominant PFGE patterns of Salmonella Typhimurium, Enteritidis and Typhi circulating in Colombia from 2005 to 2011. The figure shows the political division of Colombia and the dark grey areas indicate the departments from which the data were analysed due to presenting the highest number of isolates with predominant PFGE patterns. The boxes state the name of the department and the number of isolates shipped by the respective public health laboratory between 2005 and 2011 to the acute diarrhoeal disease (ADD) and foodborne illness (FBI) surveillance programme. The percentage was calculated on the basis of the number of total isolates recovered in the 7 years under study (n=4010). The full name of the patterns for Typhimurium is COINJPXX01.----; for Enteritidis it is COINJEGX01.----; for Typhi it is COINJPPX01.----; for Dublin it is COINJDXX01.----. See Fig. 1.
2083 isolates (51.9%) were analysed by PFGE, of which 1976 (49.2%) corresponded to the nine predominant serovars (Table 1).
Salmonella TyphimuriumThis was the main serovar recovered in the period under analysis, with a total of 1302 isolates (32.5%) (Table 1). It was mainly obtained from samples of faecal matter (73.7%) in patients of all ages, followed by blood cultures (19.5%), where 78.4% (200/255) were over 14 years of age. The remaining isolates were obtained from 10 different samples from other invasive processes.
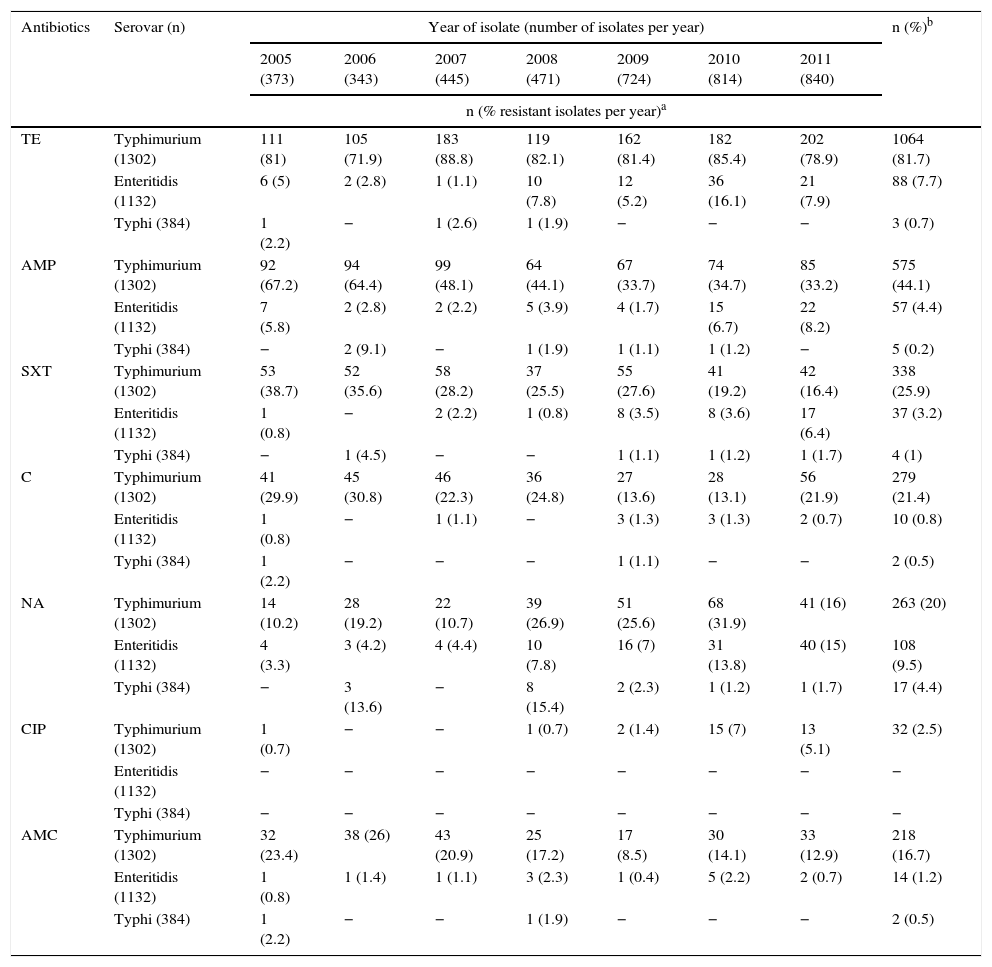
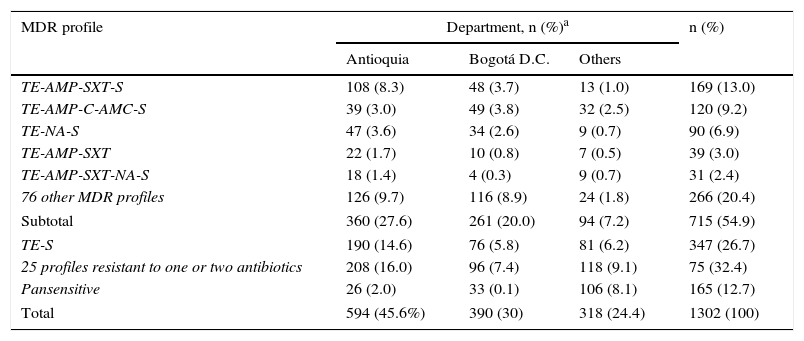
Analysing resistance in this serovar showed that 1064 isolates (82%) were resistant to TE (Table 2). Resistance to AMP, SXT, C and AMC stood at 44%, 26%, 21% and 17%, respectively, being at its lowest in the final years of the study, contrary to the pattern observed with NA resistance, which increased from 10% in 2005 to 32% in 2010, before decreasing again to 16% in 2011. Meanwhile, CIP resistance was determined in 2.5% of the isolates. For third-generation cephalosporins, a resistance of 2% was seen. MDR was observed in 54.9% of the isolates, represented by 81 profiles, the most common of which was TE-AMP-SXT-S in 169 isolates (13%), with most coming from Antioquia. The second most common profile was TE-AMP-C-AMC-S, which was found in 120 isolates (9.2%), mainly from Antioquia and Bogotá. 12.7% of the isolates were pansensitive (Table 3).
Percentage of resistance per year in isolates of Salmonella Typhimurium, Enteritidis and Typhi recovered in Colombia from 2005 to 2011.
| Antibiotics | Serovar (n) | Year of isolate (number of isolates per year) | n (%)b | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2005 (373) | 2006 (343) | 2007 (445) | 2008 (471) | 2009 (724) | 2010 (814) | 2011 (840) | |||
| n (% resistant isolates per year)a | |||||||||
| TE | Typhimurium (1302) | 111 (81) | 105 (71.9) | 183 (88.8) | 119 (82.1) | 162 (81.4) | 182 (85.4) | 202 (78.9) | 1064 (81.7) |
| Enteritidis (1132) | 6 (5) | 2 (2.8) | 1 (1.1) | 10 (7.8) | 12 (5.2) | 36 (16.1) | 21 (7.9) | 88 (7.7) | |
| Typhi (384) | 1 (2.2) | − | 1 (2.6) | 1 (1.9) | − | − | − | 3 (0.7) | |
| AMP | Typhimurium (1302) | 92 (67.2) | 94 (64.4) | 99 (48.1) | 64 (44.1) | 67 (33.7) | 74 (34.7) | 85 (33.2) | 575 (44.1) |
| Enteritidis (1132) | 7 (5.8) | 2 (2.8) | 2 (2.2) | 5 (3.9) | 4 (1.7) | 15 (6.7) | 22 (8.2) | 57 (4.4) | |
| Typhi (384) | − | 2 (9.1) | − | 1 (1.9) | 1 (1.1) | 1 (1.2) | − | 5 (0.2) | |
| SXT | Typhimurium (1302) | 53 (38.7) | 52 (35.6) | 58 (28.2) | 37 (25.5) | 55 (27.6) | 41 (19.2) | 42 (16.4) | 338 (25.9) |
| Enteritidis (1132) | 1 (0.8) | − | 2 (2.2) | 1 (0.8) | 8 (3.5) | 8 (3.6) | 17 (6.4) | 37 (3.2) | |
| Typhi (384) | − | 1 (4.5) | − | − | 1 (1.1) | 1 (1.2) | 1 (1.7) | 4 (1) | |
| C | Typhimurium (1302) | 41 (29.9) | 45 (30.8) | 46 (22.3) | 36 (24.8) | 27 (13.6) | 28 (13.1) | 56 (21.9) | 279 (21.4) |
| Enteritidis (1132) | 1 (0.8) | − | 1 (1.1) | − | 3 (1.3) | 3 (1.3) | 2 (0.7) | 10 (0.8) | |
| Typhi (384) | 1 (2.2) | − | − | − | 1 (1.1) | − | − | 2 (0.5) | |
| NA | Typhimurium (1302) | 14 (10.2) | 28 (19.2) | 22 (10.7) | 39 (26.9) | 51 (25.6) | 68 (31.9) | 41 (16) | 263 (20) |
| Enteritidis (1132) | 4 (3.3) | 3 (4.2) | 4 (4.4) | 10 (7.8) | 16 (7) | 31 (13.8) | 40 (15) | 108 (9.5) | |
| Typhi (384) | − | 3 (13.6) | − | 8 (15.4) | 2 (2.3) | 1 (1.2) | 1 (1.7) | 17 (4.4) | |
| CIP | Typhimurium (1302) | 1 (0.7) | − | − | 1 (0.7) | 2 (1.4) | 15 (7) | 13 (5.1) | 32 (2.5) |
| Enteritidis (1132) | − | − | − | − | − | − | − | − | |
| Typhi (384) | − | − | − | − | − | − | − | − | |
| AMC | Typhimurium (1302) | 32 (23.4) | 38 (26) | 43 (20.9) | 25 (17.2) | 17 (8.5) | 30 (14.1) | 33 (12.9) | 218 (16.7) |
| Enteritidis (1132) | 1 (0.8) | 1 (1.4) | 1 (1.1) | 3 (2.3) | 1 (0.4) | 5 (2.2) | 2 (0.7) | 14 (1.2) | |
| Typhi (384) | 1 (2.2) | − | − | 1 (1.9) | − | − | − | 2 (0.5) | |
AMC: amoxicillin/clavulanic acid; AMP: ampicillin; C: chloramphenicol; CIP: ciprofloxacin; NA: nalidixic acid; SXT: trimethoprim/sulfamethoxazole; TE: tetracycline.
For each serovar, the percentage of resistant isolates per year was calculated from the n per serovar per year, taking into account the values recorded in Table 1.
MDR profiles observed in the Salmonella Typhimurium isolates recovered in Colombia from 2005 to 2011.
| MDR profile | Department, n (%)a | n (%) | ||
|---|---|---|---|---|
| Antioquia | Bogotá D.C. | Others | ||
| TE-AMP-SXT-S | 108 (8.3) | 48 (3.7) | 13 (1.0) | 169 (13.0) |
| TE-AMP-C-AMC-S | 39 (3.0) | 49 (3.8) | 32 (2.5) | 120 (9.2) |
| TE-NA-S | 47 (3.6) | 34 (2.6) | 9 (0.7) | 90 (6.9) |
| TE-AMP-SXT | 22 (1.7) | 10 (0.8) | 7 (0.5) | 39 (3.0) |
| TE-AMP-SXT-NA-S | 18 (1.4) | 4 (0.3) | 9 (0.7) | 31 (2.4) |
| 76 other MDR profiles | 126 (9.7) | 116 (8.9) | 24 (1.8) | 266 (20.4) |
| Subtotal | 360 (27.6) | 261 (20.0) | 94 (7.2) | 715 (54.9) |
| TE-S | 190 (14.6) | 76 (5.8) | 81 (6.2) | 347 (26.7) |
| 25 profiles resistant to one or two antibiotics | 208 (16.0) | 96 (7.4) | 118 (9.1) | 75 (32.4) |
| Pansensitive | 26 (2.0) | 33 (0.1) | 106 (8.1) | 165 (12.7) |
| Total | 594 (45.6%) | 390 (30) | 318 (24.4) | 1302 (100) |
AMC: amoxicillin/clavulanic acid; AMP: ampicillin; C: chloramphenicol; NA: nalidixic acid; SXT: trimethoprim/sulfamethoxazole; TE: tetracycline.
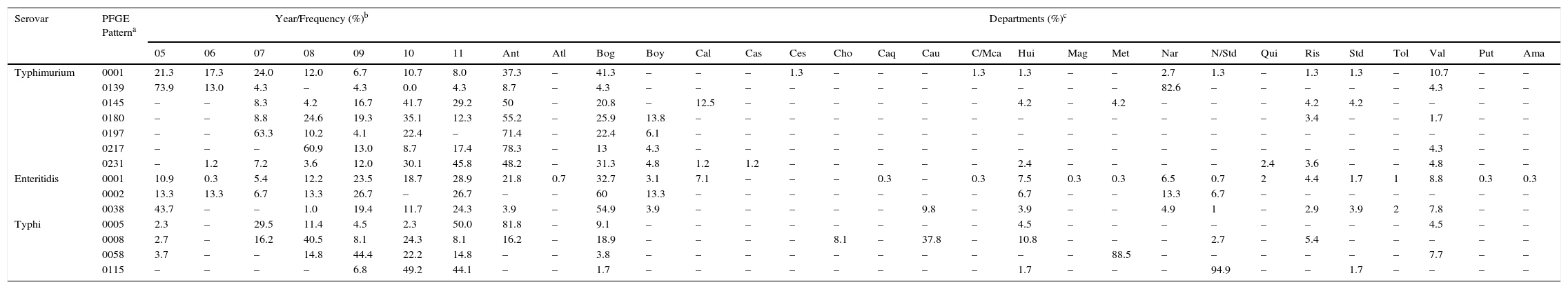
Of the 1302 isolates analysed by PFGE, 967 (74%) were processed and grouped into 439 (45% genetic variability) electrophoretic patterns (Table 1). Of these, seven patterns were observed in 34% of the isolates. The widely distributed patterns were COINJPXX01.0001 (22%), which circulated for 7 years in 10 departments, COINJPXX01.0231 (24%), which has been circulating since 2006 in nine departments, and COINJPXX01.0145 (7.2%), circulating since 2007 in seven departments (Figs. 1 and 2 and Table 4). Pattern COINJPXX01.0180 (17.4%) was mainly recovered in Antioquia and Bogotá, as well as three other departments. COINJPXX01.0139 (6.9%) and COINJPXX01.0217 (6.9%) were recovered in four departments, being predominant in Nariño and Antioquia, respectively. Pattern COINJPXX01.0197 (14.7%) was recovered in three departments (Table 4, Fig. 2).
Distribution of the predominant PFGE patterns of Salmonella Typhimurium, Enteritidis and Typhi found in Colombia from 2005 to 2011.
| Serovar | PFGE Patterna | Year/Frequency (%)b | Departments (%)c | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 05 | 06 | 07 | 08 | 09 | 10 | 11 | Ant | Atl | Bog | Boy | Cal | Cas | Ces | Cho | Caq | Cau | C/Mca | Hui | Mag | Met | Nar | N/Std | Qui | Ris | Std | Tol | Val | Put | Ama | ||
| Typhimurium | 0001 | 21.3 | 17.3 | 24.0 | 12.0 | 6.7 | 10.7 | 8.0 | 37.3 | – | 41.3 | – | – | – | 1.3 | – | – | – | 1.3 | 1.3 | – | – | 2.7 | 1.3 | – | 1.3 | 1.3 | – | 10.7 | – | – |
| 0139 | 73.9 | 13.0 | 4.3 | – | 4.3 | 0.0 | 4.3 | 8.7 | – | 4.3 | – | – | – | – | – | – | – | – | – | – | – | 82.6 | – | – | – | – | – | 4.3 | – | – | |
| 0145 | – | – | 8.3 | 4.2 | 16.7 | 41.7 | 29.2 | 50 | – | 20.8 | – | 12.5 | – | – | – | – | – | – | 4.2 | – | 4.2 | – | – | – | 4.2 | 4.2 | – | – | – | – | |
| 0180 | – | – | 8.8 | 24.6 | 19.3 | 35.1 | 12.3 | 55.2 | – | 25.9 | 13.8 | – | – | – | – | – | – | – | – | – | – | – | – | – | 3.4 | – | – | 1.7 | – | – | |
| 0197 | – | – | 63.3 | 10.2 | 4.1 | 22.4 | – | 71.4 | – | 22.4 | 6.1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| 0217 | – | – | – | 60.9 | 13.0 | 8.7 | 17.4 | 78.3 | – | 13 | 4.3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 4.3 | – | – | |
| 0231 | – | 1.2 | 7.2 | 3.6 | 12.0 | 30.1 | 45.8 | 48.2 | – | 31.3 | 4.8 | 1.2 | 1.2 | – | – | – | – | – | 2.4 | – | – | – | – | 2.4 | 3.6 | – | – | 4.8 | – | – | |
| Enteritidis | 0001 | 10.9 | 0.3 | 5.4 | 12.2 | 23.5 | 18.7 | 28.9 | 21.8 | 0.7 | 32.7 | 3.1 | 7.1 | – | – | – | 0.3 | – | 0.3 | 7.5 | 0.3 | 0.3 | 6.5 | 0.7 | 2 | 4.4 | 1.7 | 1 | 8.8 | 0.3 | 0.3 |
| 0002 | 13.3 | 13.3 | 6.7 | 13.3 | 26.7 | – | 26.7 | – | – | 60 | 13.3 | – | – | – | – | – | – | – | 6.7 | – | – | 13.3 | 6.7 | – | – | – | – | – | – | – | |
| 0038 | 43.7 | – | – | 1.0 | 19.4 | 11.7 | 24.3 | 3.9 | – | 54.9 | 3.9 | – | – | – | – | – | 9.8 | – | 3.9 | – | – | 4.9 | 1 | – | 2.9 | 3.9 | 2 | 7.8 | – | – | |
| Typhi | 0005 | 2.3 | – | 29.5 | 11.4 | 4.5 | 2.3 | 50.0 | 81.8 | – | 9.1 | – | – | – | – | – | – | – | – | 4.5 | – | – | – | – | – | – | – | – | 4.5 | – | – |
| 0008 | 2.7 | – | 16.2 | 40.5 | 8.1 | 24.3 | 8.1 | 16.2 | – | 18.9 | – | – | – | – | 8.1 | – | 37.8 | – | 10.8 | – | – | – | 2.7 | – | 5.4 | – | – | – | – | – | |
| 0058 | 3.7 | – | – | 14.8 | 44.4 | 22.2 | 14.8 | – | – | 3.8 | – | – | – | – | – | – | – | – | – | – | 88.5 | – | – | – | – | – | – | 7.7 | – | – | |
| 0115 | – | – | – | – | 6.8 | 49.2 | 44.1 | – | – | 1.7 | – | – | – | – | – | – | – | – | 1.7 | – | – | – | 94.9 | – | – | 1.7 | – | – | – | – | |
The following were calculated: n found per year of each pattern on the basis of the total n of each pattern found over the 7 years under study.
Departments where the predominant PFGE patterns were recovered. Ant: Antioquia, Atl: Atlántico, Bog: Bogotá, Boy: Boyacá, Cal: Caldas, Cas: Casanare, Ces: Cesar, Cho: Choco, Caq: Caquetá, Cau: Cauca, C/Mca: Cundinamarca, Hui: Huila, Mag: Magdalena, Met: Meta, Nar: Nariño, N/Std: North Santander, Qui: Quindío, Ris: Risaralda, Std: Santander, Tol: Tolima, Val: Valle del Cauca, Put: Putumayo, Ama: Amazonas. See Figs. 1 and 2.
Generally speaking, among the seven most common PFGE patterns for this serovar, two resistance profiles can be observed. The MDR TE-AMP-C-AMC-S profile was found in 50% (38/75) of the COINJPXX01.0001 pattern isolates. The TE and S resistance profile was predominant in five of the seven patterns (Fig. 1).
Comparing these patterns with the RDB found a 100% likeness between COINJPXX01.0139 and the ALJPXX01.0409 Argentinian pattern, as well as between COINJPXX01.0197 and the ALJPXX01.0218 Brazilian pattern, and COINJPXX01.0231 and the ALJPXX01.0326 Chilean pattern (Table 3).
Salmonella EnteritidisThis serovar had the second highest number of isolates in the 7 years under study (1132; 28.2%), overtaking the serovar Typhimurium from 2009 (Table 1). It was primarily recovered from faecal matter (66.8) in an equal percentage of adults and children, while isolates from blood cultures (26.7%) mostly came from patients over 14 years of age (69%).
TE, AMP, SXT, C and AMC resistance remained below 9%. The only antibiotic for which an increase in resistance was observed was NA: from 3% in 2005 to 15% in 2011 (Table 2). All of the isolates were sensitive to CIP and third-generation cephalosporins.
Of 1132 isolates, 523 were analysed by PFGE (46%), in which 130 (25% genetic variability) patterns were identified (Table 1); of these, three made up 77% of the isolates and were mainly recovered in Bogotá. The main pattern was COINJEGX01.0001 (56%), recovered in 19 departments over the 7 years under study; the second was COINJEGX01.038 (18%), recovered in 12 departments, and the third was COINJEGX01.002 (2.8%), mainly recovered in Bogotá and four other departments (Table 4, Fig. 2). In these three PFGE patterns, sensitive isolates were predominant (Fig. 1).
The comparison with the RDB showed that COINJEGX01.0001 was 100% identical to the ALJEGX01.0010 pattern reported in Brazil and Paraguay, and that COINJEGX01.0038 matched ALJEGX01.0001, the regional pattern for Latin America (Table 3).
Salmonella TyphiThis was the third serovar found in surveillance, with 384 isolates (9.6%). 82% was recovered from blood cultures, where those over 14 years of age represented 71.2% of the patients. 13.8% was recovered In faecal matter, where patients over 14 years of age represented 60.5%.
From 2008 to 2010, an increase in the number of Typhi isolates (Table 1) was observed in the departments of Meta and North Santander.
93.4% of the isolates were pansensitive, NA resistance was observed in 4%, and the highest resistance percentages (14% and 15%) were seen in 2006 and 2008 (Table 2).
314 isolates (89%) out of 384 were analysed by PFGE and grouped into 133 electrophoretic patterns (42% genetic variability) (Table 1). Of these, four patterns made up 45% of the isolates and were constant over the 7 years under study (Table 4). The most common pattern was COINJPPX01.0115 (18.8%), 95% of which was recovered from 2009 to 2011 in North Santander (Figs. 1 and 2 and Table 4). In second place is the COINJPPX01.0008 pattern (11.8%), distributed across seven departments. Pattern COINJPPX01.0058 (8.2%) was recovered primarily in Meta as well as Bogotá and Valle, while pattern COINJPPX01.0005 was mostly found in Antioquia, followed by Bogotá, Huila and Valle (Table 4, Fig. 2).
The comparison with the RDB showed a 100% likeness between COINJPPX01.0005 and the ALJPPX01.0016 pattern, as well as between COINJPPX01.0058 and the ALJPPX01.0048 pattern, both reported in Argentina and Chile (Table 3).
Serovars Dublin, Panama, Derby, Braenderup, Saintpaul and UgandaThese serovars occupy fourth to ninth place as regards the number of isolates (Table 1). They were mainly recovered from faecal matter (>74%), with the exception of Dublin, 72% of which was found in blood cultures. Although the number of isolates for each of the serovars remains constant over the years, the increase observed in serovar Derby for 2009 (Table 1) is striking. This was related to an outbreak in the city of Barranquilla in the Atlántico department (unpublished data), as well as the increase in serovar Panama isolates in 2010 (Table 1), where although unconnected to the outbreak, 81.8% (63/77) came from the Antioquia department from 19 different laboratories. Generally speaking, the highest resistance observed was to TE and NA. For TE, serovars Derby and Uganda were most resistant, at 87.5% and 56.5%, respectively. For NA, serovar Panama was most resistant, with 41%, followed by Braenderup, with 34%. CIP resistance was only observed in serovar Braenderup, at 10% (Table 2). Between 2 and 20 PFGE patterns were determined in these six serovars (Fig. 1, Table 1). On comparing the predominant patterns of each of these serovars with the RDB, it was found that only the COINJDXX01.0001 pattern of serovar Dublin, recovered in various departments (Fig. 2) had a 100% likeness with the ALJDXX01.0001 pattern recovered in Argentina and Guatemala (Table 3).
Other serovarsThe remaining serovars (n=83) were mainly recovered from faecal matter in 74.1% and blood cultures in 14.8%. In the population over 6 years of age, 11.1% of the isolates came from urine (4.5%), secretions (1.2%), bodily fluid (0.7%), cerebrospinal fluid (CSF) (0.6%), abscesses (0.4%), biopsies and wounds (0.3%), tracheal aspirate, vomit and tissue (0.2%) and unrecorded sources (2.5%). Of these, the serovar Javiana was most frequently recovered from CSF (5.8%). No predominant resistance profile was found. 25 serovars related to outbreaks of FBI or extraintestinal disease were analysed by PFGE (Table 1).
DiscussionAnalysing the data obtained from the 2005 to 2011 surveillance showed that the main salmonellosis-causing serovars in Colombia are Typhimurium, Enteritidis and Typhi, thus confirming reports from previous years.13,15,16 In this study, a serovar displacement was observed from 2009 to 2011, when Enteritidis, with a wider national distribution, overtook Typhimurium. Of the 32 Colombian departments and the Capital District of Bogotá that participate in surveillance, Antioquia and Bogotá provided the highest number of isolates (Fig. 2).
Typhimurium and Enteritidis were mainly recovered from faecal matter in symptoms of gastroenteritis, thus proving the capacity of these serovars to cause diarrhoea.17,18 Globally, Enteritidis is the main serovar in Europe, Asia and Latin America, while Typhimurium is predominant in North America and the Oceania region.14,19 In Latin America, an increase in Typhimurium has been observed, from 14.5% in 2001 to 24% in 2007.19 Infection by these two serovars is mainly attributed to a lack of hygiene when preparing foods such as meat and vegetables in the case of Typhimurium, and undercooking contaminated eggs and roast chicken for Enteritidis, making them a major public health problem.3,20–23 Added to this problem is the MDR reported for serovar Typhimurium worldwide, which has been associated with its capacity to cause invasive disease in humans.3 In this study, we identify various MDR profiles in Typhimurium, but none associated with any of the predominant PFGE patterns, suggesting that MDR dissemination in this serovar does not occur by clonal dispersal, but possibly by means of mobile genetic elements.
Historically, Typhimurium was the first Salmonella serovar to acquire MDR to five different families of antibiotics, which was reported in the 1960s in the United Kingdom, associated with livestock and cattle production, increasing spectacularly in the 1990s, while reports of MDR for Enteritidis remained below 1%.24 In general, this tendency has been maintained ever since, making Typhimurium one of the main Salmonella serovars associated with MDR worldwide, while the MDR levels for Enteritidis remain low. In the United States in 2013, Typhimurium was reported as one of the predominant serovars with MDR to 4, 5 and 7 different antibiotics, while Enteritidis was the most common serovar in nalidixic acid-resistant isolates.25 In Colombia, differences in resistance levels between these two serovars, as well as being intrinsic to each of them, may be due to the use of antibiotics as promoters of growth in pig and poultry production within the country. Local studies have documented the presence of Salmonella spp. in the pre-slaughter and slaughter stages of pig production, where over 90% are resistant to two or more antibiotics.26,27 The serovar Enteritidis has been reported in raw and ready-for-sale chicken, but with low resistance levels.28,29 The serovar Typhimurium, which carries genes resistant to between one and five antibiotics, was found in processed food samples at points of sale.30 These studies suggest an association between the levels of resistance observed in serovars Typhimurium and Enteritidis, recovered from both clinical samples and some foods. However, in Colombia, further studies are needed to allow us to establish a link between Salmonella serovars circulating in animals and their resistance and direct relationship with the isolates recovered in clinical samples. The presence of multiple genes that confer MDR to serovar Paratyphi B isolates of the Java and Heidelberg variety in poultry production is also worrying as, despite not being clinically relevant serovars in Colombia, they may be behind the dissemination to the main serovars recovered in human clinical samples.28,31
In this study, serotyping with PFGE reaffirmed the genetic diversity of the serovar Typhimurium, as well as the homogeneity of Enteritidis observed previously in Colombian isolates.13,32 As regards Typhimurium, the COINJPXX01.0001 electrophoretic pattern remained predominant in Colombia from 1998 to 2011, except for the period between 2002 and 2004, when it was replaced by COINJPXX01.0062.13 With respect to the other patterns, a shift in the predominant ones was observed in comparison to those reported between 1997 and 2004, thus reaffirming their genetic plasticity. This result is important because it generated different regional PFGE patterns to those reported, which suggests an increase in common contamination sources, as well as the emergence of clones with a high dissemination potential in the region.14 For Enteritidis, two of the three predominant PFGE patterns in Colombia circulate in the region. One of them matches the pattern prevalent in Latin America (ALJEGX01.0001), while pattern ALJEGX01.0010 has been reported in Brazil and Paraguay. These results suggest the dissemination of new Enteritidis strains in the region.14
Typhi is the third most prevalent serovar in the country, suggesting that typhoid fever continues to be a public health problem for Colombia, despite being considered a disease of low endemicity.33 Globally, it is estimated that typhoid fever causes around 11.9 million cases and approximately 129,000 deaths associated with contaminated water consumption.34 In Colombia, 104 cases of typhoid and paratyphoid fever were reported to the National Surveillance System in 2011, of which 93% (n=96) were laboratory-confirmed.33 Isolates of this serovar continue to be pansensitive in the country, which proves advantageous in the treatment of typhoid fever. Using PFGE, it has been determined that Typhi isolates recovered in Colombia are genetically heterogeneous despite some patterns being related to outbreaks.14,35 The circulation of two patterns shared with Argentina (ALJPPX01.0002 and ALJPPX01.0013)35 had previously been reported, and for this study two new patterns were identified (ALJPPX01.0016 and ALJPPX01.0048), shared with Argentina and Chile. These results suggest the dissemination of Typhi strains within the continent, possibly through travellers.
The different non-typhoidal serovars (NTS) recovered most frequently in clinical samples are potentially a reflection of the serovars circulating in foods for human consumption. The nine predominant serovars in Colombia differ from those reported in Europe and the United States. It has been suggested that changes in the frequency of specific serovars may be the result of movements of people, animals and foods; for this reason, the constant surveillance of this pathogen in communities is important.36 These serovars are resistant to one, two or multiple antibiotics, with TE and NA being the antibiotics to which they present the highest resistance. The serovars with high rates of resistance to TE are those associated with cattle, pig and poultry production (Typhimurium, Derby, Saintpaul and Uganda), thus suggesting an uncontrolled use of this antibiotic in food production,23,37–39 although in Colombia we have no direct evidence of its indiscriminate use. The resistance recorded for NA makes us aware of the possible trend of fluoroquinolone resistance, particularly in serovars Typhimurium, Panama, Braenderup and Uganda, despite the sensitivity observed for CIP. This result is probably down to the fact that it was evaluated with cut-off points established before 2012 by the CLSI, which were very high for CIP.40 Moreover, it is important to highlight that a TE and NA resistance of over 60% has recently been reported in Salmonella spp. isolates recovered from chicken in Colombia, which could be related to the resistance observed in human isolates analysed in this study.28 The percentages of third-generation cephalosporin resistance are below 1%, thus representing a treatment advantage in the presence of invasive salmonellosis symptoms, contrary to what has been observed in several other countries.41,42
It is important to mention that NTS are also recovered from other samples associated with invasive symptoms, such as blood cultures, CSF and urine. Of these, serovar Dublin is striking as it was recovered in blood cultures after the serovar Typhi, in accordance with that reported in the literature regarding its capacity to generate invasive symptoms in humans.43 Changes in the epidemiology of some of these serovars have been reported, where clonal lineages generate invasive symptoms.44 Although in Colombia we do not know whether there is a link between a specific lineage and invasive cases caused by NTS, the results suggest an increase in these types of isolates, as reported in Europe and the United States; it is therefore important to continue monitoring the behaviour of these serovars nationwide.45
LimitationsThe surveillance process may lead to under-reporting in the system. However, in this report an increase in laboratory notifications and shipments of Salmonella spp. isolates to the NIH Microbiology Group was seen in the years under analysis.
ConclusionsAnalysing the Salmonella spp. isolates recovered in the 2005–2011 surveillance programme confirmed that serovars Typhimurium, Enteritidis and Typhi continue to be the main causes of salmonellosis in Colombia. We managed to establish the circulation of predominant PFGE patterns for each serovar. Generally speaking, in Colombia Salmonella spp. presents high percentages of TE and NA resistance, but continues to be sensitive to CIP and third-generation cephalosporins. Surveillance of this enteropathogen allows us to understand the dynamics of the Salmonella spp. serovars and to identify endemic clones in Colombia with a view to laying foundations for optimal treatment in infections generated by this pathogen and being able to design programmes for reducing the spread of MDR strains.
FundingMicrobiology Group, Directorate of Public Health Networks and the National Laboratory Network and Directorate of Research, National Institute of Health, Colombia.
Conflicts of interestThe authors state that they have no conflicts of interest. The people mentioned in the Acknowledgements section are aware of the contents of this article and consent to appearing in it.
Thank you to all of the Colombian PHLs, for actively taking part in the ADD surveillance programme. To Dr Elizabeth Castañeda, Emeritus Research Fellow of the Colombian National Institute of Health, for her critical review and contributions to the manuscript. To the PulseNet-LA and Caribbean Network, for comparing our patterns to the RDB, as well as to the people in charge of the network in the different countries across Latin America. To Enrique Perez Gutierrez, PAHO/WHO, of the Communicable Diseases and Health Analysis Department, for supporting the PulseNet-LA and Caribbean network.
Please cite this article as: Rodríguez EC, Díaz-Guevara P, Moreno J, Bautista A, Montaño L, Realpe ME, et al. Vigilancia por laboratorio de Salmonella enterica en casos clínicos humanos en Colombia 2005 a 2011. Enferm Infecc Microbiol Clin. 2017;35:417–425.