The 13-valent pneumococcal conjugate vaccine (PCV13) universal vaccination programme was introduced in December 2016 in Andalusia.
MethodsA cross-sectional study was conducted on the molecular epidemiology of pneumococcal nasopharyngeal colonization. A total of 397 healthy children were recruited from primary healthcare centres in Seville for the periods 1/4/2018 to 28/2/2020 and 1/11/2021 to 28/2/2022 (PCV13 period). Data from a previous carriage study conducted among healthy and sick children from 1/01/2006 to 30/06/2008 (PCV7 period), were used for comparison of serotype/genotype distributions and antibiotic resistance rates.
ResultsOverall, 76 (19%) children were colonized with S. pneumoniae during the PCV13 period and there were information available from 154 isolates collected during the PCV7 period. Colonization with PCV13 serotypes declined significantly in the PCV13 period compared with historical controls (11% vs 38%, p = 0.0001), being serotypes 19F (8%), 3 (1%) and 6B (1%) the only circulating vaccine types. Serotypes 15B/C and 11A were the most frequently identified non-PCV13 serotypes during the PCV13 period (14% and 11%, respectively); the later one increased significantly between time periods (p = 0.04). Serotype 11A was exclusively associated in the PCV13 period with ampicillin-resistant variants of the Spain9V-ST156 clone (ST6521 and genetically related ST14698), not detected in the preceding period.
ConclusionsThere was a residual circulation of vaccine types following PCV13 introduction, apart from serotype 19F. Serotype 11A increased between PCV13 and PCV7 periods due to emergence and clonal expansion of ampicillin-resistant genotype ST6521.
El programa de vacunación universal con la vacuna antineumocócica conjugada 13-valente (VNC13) se implantó en Andalucía en diciembre de 2016.
MétodosEstudio transversal de colonización nasofaríngea por Streptococcus pneumoniae. Se reclutaron 397 niños sanos en centros de atención primaria de Sevilla durante los periodos 1/4/2018 al 28/2/2020 y 1/11/2021 al 28/2/2022 (periodo VNC13). Se utilizó una colección histórica de un estudio de colonización desarrollado en niños sanos y con infección respiratoria superior entre el 1/01/2006 y 30/06/2008 (período VNC7) para comparar las distribuciones de serotipos/genotipos y las tasas de resistencias antibióticas.
ResultadosUn total de 76 (19%) niños estaban colonizados con S. pneumoniae en el período VNC13 y se dispuso de 154 aislamientos del período VNC7. La colonización por serotipos incluidos en VNC13 disminuyó significativamente entre los periodos VNC13 y VNC7 (11% vs 38%, p = 0,0001), siendo los serotipos 19F (8%), 3 (1%) y 6B (1%) los únicos serotipos vacunales circulantes. Los serotipos 15B/C y 11A fueron los serotipos no VNC13 más prevalentes durante el período VNC13 (14% y 11%, respectivamente); este último se incrementó significativamente entre periodos de tiempo (p = 0,04). El serotipo 11A se asoció exclusivamente en el periodo VNC13 con variantes resistentes a la ampicilina del clon Spain9V-ST156 (ST6521 y genéticamente relacionado ST14698), no detectadas en el periodo anterior.
ConclusionesHubo una circulación muy residual de los serotipos vacunales durante el periodo VNC13, con excepción del serotipo 19F. El serotipo 11A se incrementó significativamente entre los periodos VNC13 y VNC7 por expansión clonal del genotipo resistente a la ampicilina ST6521.
Streptococcus pneumoniae is a common commensal of the human nasopharynx. Pneumococcal colonisation is a universal process, with a higher prevalence in young children, and is considered a requirement for the development of pneumococcal disease (mucosal or invasive).1 This is highly relevant in infectious disease throughout the world due to its prevalence and high disease burden.2
Pneumococcal conjugate vaccines (PCVs) have significantly reduced the incidence of invasive pneumococcal disease (IPD) in the immunised population and also, although to a lesser extent, among non-immunised people due to the “herd effect”.3,4 However, its effectiveness is limited by the partial coverage of serotypes and may decrease over time due to the so-called replacement phenomenon (increase in the incidence of IPD due to non-vaccine serotypes [NVS] resulting from their greater presence in nasopharyngeal colonisation due to vaccine immunity pressure on vaccine serotypes [VS]).5 Active surveillance of the epidemiology of IPD after the introduction of PCVs is essential to analyse the impact of vaccination on the incidence of IPD in all age groups, changes in the distribution of serotypes and the possible appearance of serotype replacement. As a complement to epidemiological surveillance in IPD, cross-sectional studies of nasopharyngeal carriage are of great importance to understand the underlying mechanisms of the "herd effect" and serotype replacement, and they inform about changes in prevalence, resistance rates and the invasive potential of replacement NVS in the nasopharynx.
Systematic vaccination against pneumococcus with 13-valent PCV (PCV13) was introduced in December 2016 in the childhood vaccination schedule of Andalusia (Spain). Various PCV formulations had already been available (PCV7, PCV10 and PCV13 since 2001, 2009 and 2010, respectively) on the private market with variable anti-pneumococcal vaccination rates (41–61%) among the Andalusian paediatric population.6,7 The PCV13 vaccination coverage in the Andalusian paediatric population after the start of the childhood vaccination programme ranged from 97.2% in 2018 to 98.2% in 2020 (Moreno D. personal communication).
Our group has conducted two parallel studies of molecular epidemiological surveillance of IPD and colonisation of the nasopharynx in the Andalusian paediatric population for a better understanding of the benefits of the systematic vaccination programme against pneumococcus and to monitor the evolution of NVS. The objective of this study was to describe the distribution of serotypes and genotypes and the rates of antimicrobial resistance in pneumococcal isolates obtained from the nasopharynx of healthy children under five years of age treated at various health centres in the city of Seville after the introduction of universal childhood vaccination with PCV13. In addition, and as a secondary objective, this information was compared with other similar information obtained in the last 30 months from a cross-sectional study of nasopharyngeal carriage conducted in the same geographical area in a period with partial vaccination with 7-valent PCV (PCV7).7,8
MethodsDesign, scope and study periodThis was a cross-sectional, observational descriptive study on the epidemiology of pneumococcal nasopharyngeal colonisation. A total of 394 healthy children between the ages of six months and five years, seen at a well-child visit or on demand in 10 public and private primary care centres in the city of Seville, were selected by logistical criteria. The study covered the periods 01/04/2018−28/02/2020 and 01/11/2021−28/02/2022 (PCV13 period).
Sampling and data collection proceduresParticipating children were selected without prior randomisation criteria. This study was conducted taking into account current legislation, the ethical standards of the Declaration of Helsinki and the guidelines for good clinical laboratory practice. Parents or legal guardians were informed verbally and in writing of the characteristics of the study and they were asked to sign the informed consent form. The study was approved by the Ethics Committee of the Hospital Universitario Virgen del Rocío [Virgen del Rocío University Hospital] in Seville (code 2017/176).
Prior to obtaining the sample from the nasopharynx, information was collected on the following potential risk factors for pneumococcal colonisation: age, gender, nursery attendance, use of antibiotics in the previous two months and type of antibiotic used, passive smoking and vaccination status against pneumococcus. The samples from the nasopharynx were obtained by healthcare professionals previously trained in the procedure according to a previously described methodology.9 The samples were transported to the Microbiology Department of the Virgen del Rocío University Hospital within 24 h for conventional microbiological processing, after which they were frozen at –80 °C until the serotyping and genotyping procedures were later completed.
Microbiological studies, serotyping and genotypingNasopharyngeal samples were plated on Columbia blood agar plates, supplemented with 5 μg/ml gentamicin, and α-haemolytic colonies were identified as pneumococci based on their morphology, optochin susceptibility and bile solubility. For serotyping and genotyping, genomic DNA was extracted from the pneumococcal isolates and their complete sequencing was carried out at the Instituto de Biomedicina de Sevilla [Seville Institute of Biomedicine]. Determination of the capsular serotype was performed by bioinformatics analysis of the genomic sequences of the capsular locus.10 The VS group included all serotypes covered by the PCV13 formulation (1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, 23F). The remaining serotypes were considered to be NVS. Genotyping was performed by multilocus sequence typing according to standard methodology.11 A clonal complex (CC) was defined as a sequence type (ST) sharing at least five of the seven allelic variants.
Antimicrobial susceptibility to penicillin, ampicillin, cefotaxime and erythromycin was determined by E-test using the breakpoints recommended by EUCAST in 2022 for assigning interpretive categories (https://www.eucast.org/clinical_breakpoints). Within the non-susceptible category, resistant strains and those sensitive to high doses were combined.
Historical PCV7 controlsTo compare the distribution of serotypes and genotypes and the rates of antibiotic resistance, a historical collection of 154 pneumococcal isolates from the PCV7 time period between 01/01/2006 and 30/06/2008, which had been partially reported (for the period 01/01/2006 to 30/06/2006) in the literature, were used.8 These colonising strains were obtained in a study of nasopharyngeal carriage conducted at four health centres and two hospital accident and emergency departments in Seville, according to the described methodology.7 In this period, antimicrobial susceptibility to oral amoxicillin was evaluated, but not to ampicillin, as in the PCV13 period.
Statistical analysisStatistical processing was performed using the SSPS 26.0 package. Categorical variables were compared using the χ2 and Fisher's exact tests, as appropriate. Variables with a p-value <0.10 in the univariate analysis of predictors of pneumococcal carriage status were entered into a binary logistic regression model. It was considered significant in a test when the p-value was <0.05.
ResultsDemographic characteristicsA total of 76 (19%) nasopharyngeal colonising strains of S. pneumoniae were identified in 397 healthy children during the PCV13 period, in addition to a historical collection of 154 (35%) pneumococcal nasopharyngeal isolates obtained from 443 participants (89 healthy children [20%] and 354 children with mild upper respiratory tract infections [80%]) during the PCV7 period.
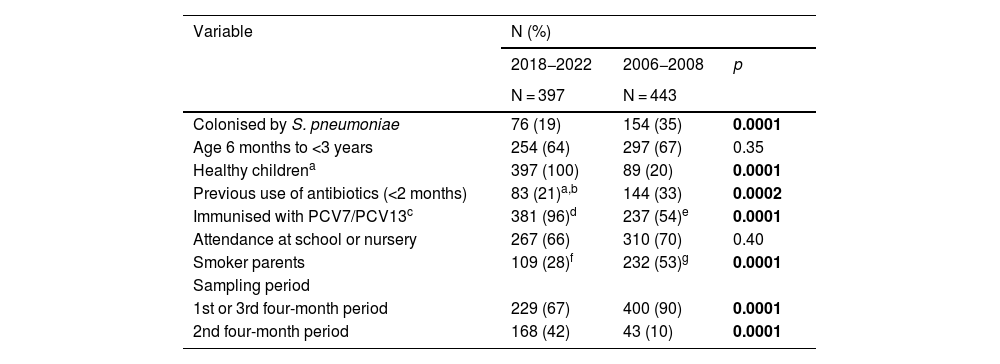
Compared to the PCV7 period, the PCV13 period was associated with lower rates of colonisation (19% vs 35%; p = 0.0001), antibiotic use in the previous two months (21% vs 33%; p = 0.0002) and smoking in one of the parents (28% vs 53%; p = 0.0001), and with a higher proportion of healthy children (100% vs 20%; p = 0.0001) and children immunised with PCV (96% vs 54%; p = 0.0001) (Table 1). On the other hand, there were no differences between the two periods in the proportions of younger children (six months to three years) and those attending a nursery or primary school. Finally, the recruitment of participants was significantly higher in the second four-month period in the contemporary study than in the historical one (42% vs 10%; p = 0.0001).
Comparative analysis of the demographic variables in the nasopharyngeal colonisation studies of the PCV13 and PCV7 periods.
| Variable | N (%) | ||
|---|---|---|---|
| 2018−2022 | 2006−2008 | p | |
| N = 397 | N = 443 | ||
| Colonised by S. pneumoniae | 76 (19) | 154 (35) | 0.0001 |
| Age 6 months to <3 years | 254 (64) | 297 (67) | 0.35 |
| Healthy childrena | 397 (100) | 89 (20) | 0.0001 |
| Previous use of antibiotics (<2 months) | 83 (21)a,b | 144 (33) | 0.0002 |
| Immunised with PCV7/PCV13c | 381 (96)d | 237 (54)e | 0.0001 |
| Attendance at school or nursery | 267 (66) | 310 (70) | 0.40 |
| Smoker parents | 109 (28)f | 232 (53)g | 0.0001 |
| Sampling period | |||
| 1st or 3rd four-month period | 229 (67) | 400 (90) | 0.0001 |
| 2nd four-month period | 168 (42) | 43 (10) | 0.0001 |
In bold, statistically significant p-values.
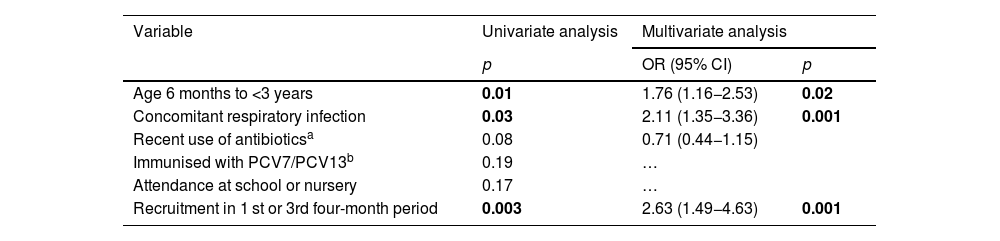
In the univariate analysis, pneumococcal colonisation in both the study periods was significantly associated with age <3 years, concomitant presence of upper respiratory tract infection (sick children) and recruitment in the first or third four-month period (Table 2). In the multivariate analysis, these variables continued to be positively associated with increased risk of colonisation.
Univariate and multivariate analysis of demographic variables associated with an increased risk of pneumococcal colonization.
| Variable | Univariate analysis | Multivariate analysis | |
|---|---|---|---|
| p | OR (95% CI) | p | |
| Age 6 months to <3 years | 0.01 | 1.76 (1.16−2.53) | 0.02 |
| Concomitant respiratory infection | 0.03 | 2.11 (1.35−3.36) | 0.001 |
| Recent use of antibioticsa | 0.08 | 0.71 (0.44−1.15) | |
| Immunised with PCV7/PCV13b | 0.19 | … | |
| Attendance at school or nursery | 0.17 | … | |
| Recruitment in 1 st or 3rd four-month period | 0.003 | 2.63 (1.49−4.63) | 0.001 |
In bold, statistically significant p-values.
In all, 27 and 32 different individual serotypes were identified in the total of 76 and 154 isolates from the contemporary and historical study periods, respectively. Colonisation by VS included in PCV13 decreased significantly between the PCV13 and PCV7 periods (n = 8, 11% vs n = 58, 38%; p = 0.0001).
During the most recent study period, only the vaccine serotypes 19F (n = 6, 8%), 3 (n = 1, 1%) and 6B (n = 1, 1%) were identified (Fig. 1). These had been in circulation in the PCV7 period, without significant differences being observed between the two periods. The remaining VS present in the historical period and eradicated in the contemporary period included the serotypes: 19A (n = 15, 10%), 6A (n = 9, 6%), 23F (n = 4, 4%), 9V (n = 3, 2%), 7F (n = 2, 1%), 14 (n = 2, 1%), 5 (n = 1, 1%) and 18C (n = 1, 1%). The proportion of serotype 19A isolates decreased significantly between the two time periods (p = 0.003).
On the other hand, the following NVS with prevalence ≥3% were identified in the PCV13 period: 15B/C (n = 11, 14%), 11A (n = 8, 11%), 23A (n = 5, 7%), 10A (n = 4, 5%), 16F (n = 4, 5%), 31 (n = 4, 5%), non-typeable (NT) (n = 4, 5%), 23B (n = 3, 4%), 21 (n = 3, 4%), 6C (n = 2, 3%), 15A (n = 2, 3%), 22F (n = 2, 3%), 34 (n = 2, 3%), 35B (n = 2, 3%), 35F (n = 2, 3%) and 38 (n = 2, 3%). Other NVS present during this time period are described in Fig. 1. When comparing the PCV13 and PCV7 periods, a significant increase in the proportions of serotypes 11A and 31 was observed (p = 0.04 for both serotypes) and there were no significant changes in the remaining NVS.
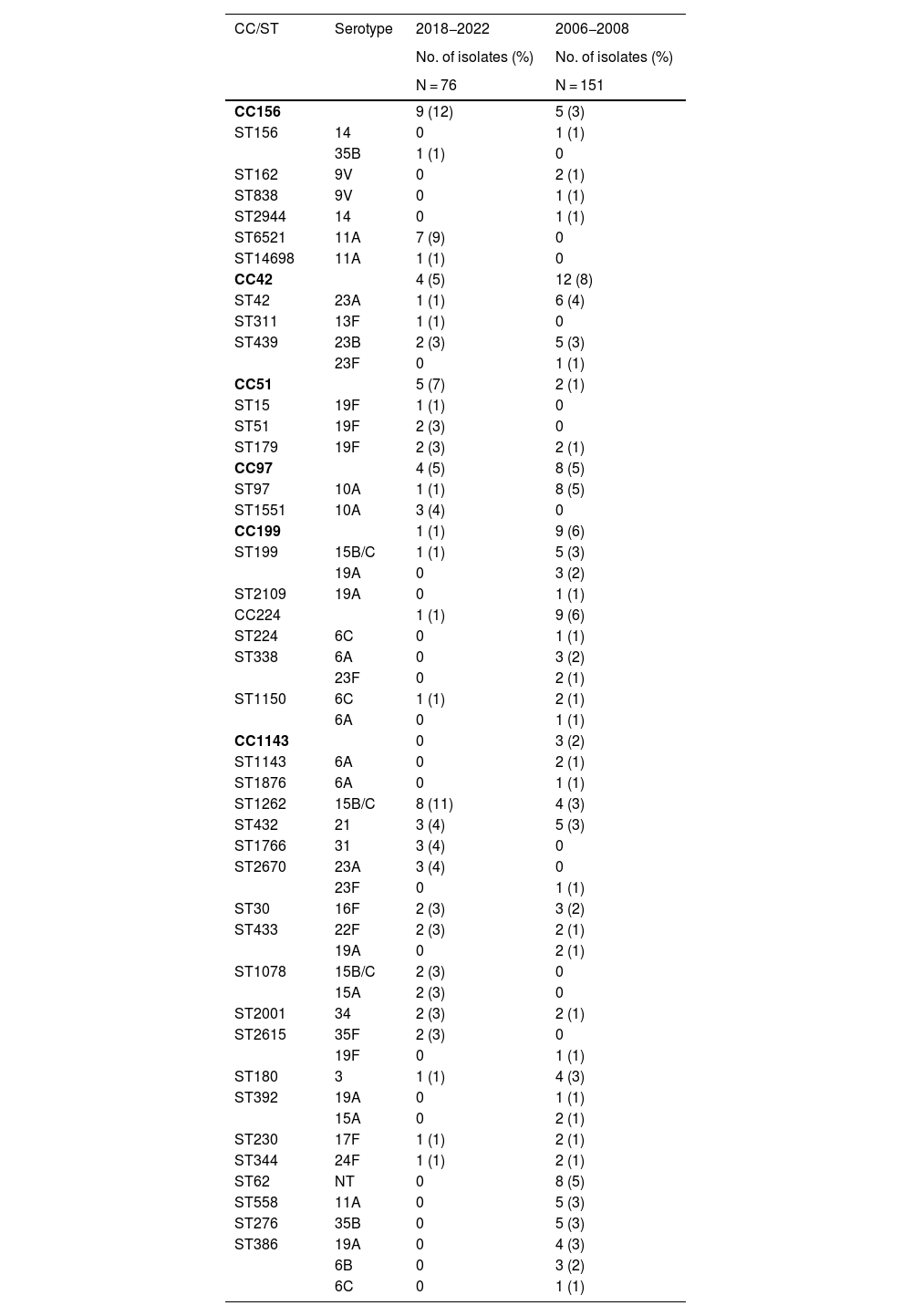
Genotype distributionA total of 40 and 67 different sequence types were identified by multilocus sequence typing, out of a total of 76 and 152 genotyped isolates from the PCV13 and PCV7 periods, respectively. The most common clonal complexes in the PCV13 period were CC156 (n = 9, 12%), followed by CC42, CC51 and CC97 (n = 4, 5%, all three), while the sequence types with the highest frequencies in this same period were ST1262 (n = 8, 11%), ST433, ST1760 and ST2670 (n = 3, 4%, all three) (Table 3). CC156 was predominantly associated in the PCV13 period with the ST6521 sequence type, a genetic variant of the global clone Spain9V-ST156 expressed as serotype 11A. This serotype was exclusively associated with clone ST62 in the historical period and was replaced by ST6521 in more recent years. ST1262 was expressed as serotype 15B/C and, unlike ST6521, remained stable during both study periods. Other genotypes identified in the historical and contemporary periods are shown in Table 2 and in the table of additional material in Appendix A.
Clonal complexes and sequence types with ≥3 isolates in the PCV13 or PCV7 study periods and associated serotypes.
| CC/ST | Serotype | 2018−2022 | 2006−2008 |
|---|---|---|---|
| No. of isolates (%) | No. of isolates (%) | ||
| N = 76 | N = 151 | ||
| CC156 | 9 (12) | 5 (3) | |
| ST156 | 14 | 0 | 1 (1) |
| 35B | 1 (1) | 0 | |
| ST162 | 9V | 0 | 2 (1) |
| ST838 | 9V | 0 | 1 (1) |
| ST2944 | 14 | 0 | 1 (1) |
| ST6521 | 11A | 7 (9) | 0 |
| ST14698 | 11A | 1 (1) | 0 |
| CC42 | 4 (5) | 12 (8) | |
| ST42 | 23A | 1 (1) | 6 (4) |
| ST311 | 13F | 1 (1) | 0 |
| ST439 | 23B | 2 (3) | 5 (3) |
| 23F | 0 | 1 (1) | |
| CC51 | 5 (7) | 2 (1) | |
| ST15 | 19F | 1 (1) | 0 |
| ST51 | 19F | 2 (3) | 0 |
| ST179 | 19F | 2 (3) | 2 (1) |
| CC97 | 4 (5) | 8 (5) | |
| ST97 | 10A | 1 (1) | 8 (5) |
| ST1551 | 10A | 3 (4) | 0 |
| CC199 | 1 (1) | 9 (6) | |
| ST199 | 15B/C | 1 (1) | 5 (3) |
| 19A | 0 | 3 (2) | |
| ST2109 | 19A | 0 | 1 (1) |
| CC224 | 1 (1) | 9 (6) | |
| ST224 | 6C | 0 | 1 (1) |
| ST338 | 6A | 0 | 3 (2) |
| 23F | 0 | 2 (1) | |
| ST1150 | 6C | 1 (1) | 2 (1) |
| 6A | 0 | 1 (1) | |
| CC1143 | 0 | 3 (2) | |
| ST1143 | 6A | 0 | 2 (1) |
| ST1876 | 6A | 0 | 1 (1) |
| ST1262 | 15B/C | 8 (11) | 4 (3) |
| ST432 | 21 | 3 (4) | 5 (3) |
| ST1766 | 31 | 3 (4) | 0 |
| ST2670 | 23A | 3 (4) | 0 |
| 23F | 0 | 1 (1) | |
| ST30 | 16F | 2 (3) | 3 (2) |
| ST433 | 22F | 2 (3) | 2 (1) |
| 19A | 0 | 2 (1) | |
| ST1078 | 15B/C | 2 (3) | 0 |
| 15A | 2 (3) | 0 | |
| ST2001 | 34 | 2 (3) | 2 (1) |
| ST2615 | 35F | 2 (3) | 0 |
| 19F | 0 | 1 (1) | |
| ST180 | 3 | 1 (1) | 4 (3) |
| ST392 | 19A | 0 | 1 (1) |
| 15A | 0 | 2 (1) | |
| ST230 | 17F | 1 (1) | 2 (1) |
| ST344 | 24F | 1 (1) | 2 (1) |
| ST62 | NT | 0 | 8 (5) |
| ST558 | 11A | 0 | 5 (3) |
| ST276 | 35B | 0 | 5 (3) |
| ST386 | 19A | 0 | 4 (3) |
| 6B | 0 | 3 (2) | |
| 6C | 0 | 1 (1) |
In bold, CC and ST identified only during the 2018−2022 period.
CC: clonal complexes; ST: sequence types.
Of the 76 strains from the PCV13 period, 20 (16%) showed reduced susceptibility to penicillin, of which two isolates (3%) were classified as resistant to penicillin. The rates of reduced susceptibility and resistance to the other two β-lactam antibiotics evaluated in both periods were as follows: ampicillin (17% and 16%) and cefotaxime (7% and 0%); 15 strains (20%) also showed resistance to erythromycin. The antimicrobial resistance rates of the PCV7 period did not show statistically significant differences compared to those of the PCV13 period for penicillin (with reduced susceptibility to penicillin [32% vs 26%; p = 0.34] and resistant to penicillin [0% vs 3%; p = 0.11]) or cefotaxime (reduced susceptibility [4% vs 7%; p = 0.37] and resistance [0% vs 0%]), but were, in contrast, significantly higher for erythromycin (38% vs 20%; p = 0.006).
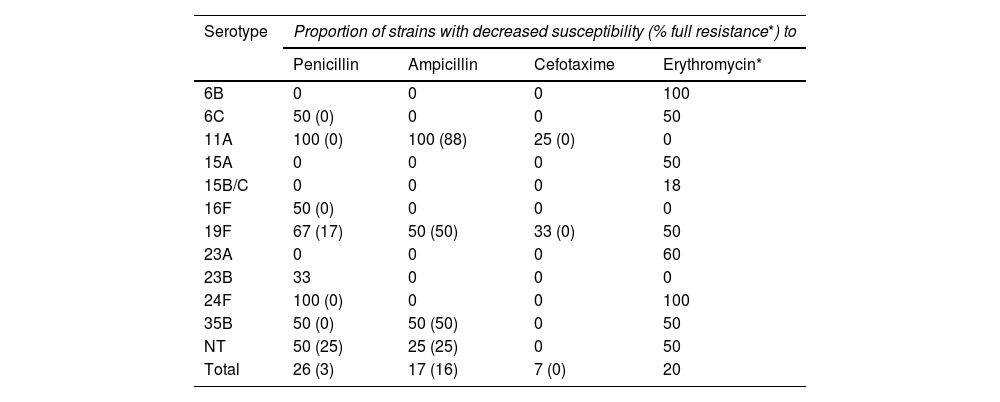
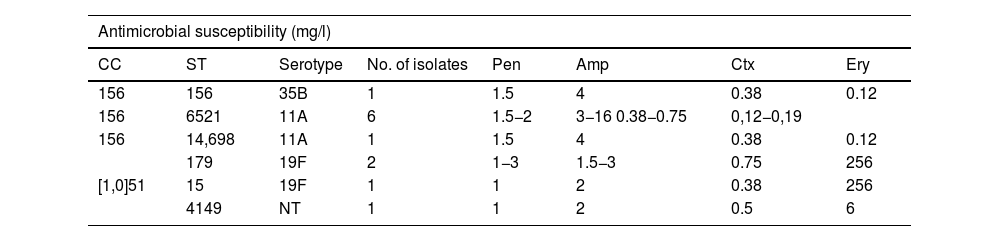
Table 4 shows the distribution in the PCV13 period of the serotypes with reduced susceptibility to β-lactam antibiotics and with resistance to erythromycin. The isolates with resistance to ampicillin in these years corresponded mostly to the serotype 11A strains (clone ST6521 and the genetically related ST14698) (8/12, 67%), while, in contrast, the five isolates of the ST62 genotype associated with this serotype in the PCV7 period showed full susceptibility to oral amoxicillin (MIC 0.06 mg/l). The remaining serotypes and genotypes with resistance to ampicillin in the PCV13 period are described in Table 5.
Serotypes associated with decreased susceptibility to β-lactam antibiotics and erythromycin in the 76 nasopharyngeal isolates from the PCV13 period.
| Serotype | Proportion of strains with decreased susceptibility (% full resistance*) to | |||
|---|---|---|---|---|
| Penicillin | Ampicillin | Cefotaxime | Erythromycin* | |
| 6B | 0 | 0 | 0 | 100 |
| 6C | 50 (0) | 0 | 0 | 50 |
| 11A | 100 (0) | 100 (88) | 25 (0) | 0 |
| 15A | 0 | 0 | 0 | 50 |
| 15B/C | 0 | 0 | 0 | 18 |
| 16F | 50 (0) | 0 | 0 | 0 |
| 19F | 67 (17) | 50 (50) | 33 (0) | 50 |
| 23A | 0 | 0 | 0 | 60 |
| 23B | 33 | 0 | 0 | 0 |
| 24F | 100 (0) | 0 | 0 | 100 |
| 35B | 50 (0) | 50 (50) | 0 | 50 |
| NT | 50 (25) | 25 (25) | 0 | 50 |
| Total | 26 (3) | 17 (16) | 7 (0) | 20 |
Serotypes and genotypes associated with resistance to ampicillin (MIC >1 μg/ml) during the 2018–2022 period.
| Antimicrobial susceptibility (mg/l) | |||||||
|---|---|---|---|---|---|---|---|
| CC | ST | Serotype | No. of isolates | Pen | Amp | Ctx | Ery |
| 156 | 156 | 35B | 1 | 1.5 | 4 | 0.38 | 0.12 |
| 156 | 6521 | 11A | 6 | 1.5−2 | 3−16 0.38−0.75 | 0,12−0,19 | |
| 156 | 14,698 | 11A | 1 | 1.5 | 4 | 0.38 | 0.12 |
| 179 | 19F | 2 | 1−3 | 1.5−3 | 0.75 | 256 | |
| [1,0]51 | 15 | 19F | 1 | 1 | 2 | 0.38 | 256 |
| 4149 | NT | 1 | 1 | 2 | 0.5 | 6 | |
Genotypes and serotypes in bold were either not detected (ST6521, ST14698, ST15 and ST4149) or were not associated with amoxicillin resistance (CC51, ST179) during the PCV7 period.
Amp: ampicillin; CC: clonal complex; Ctx: cefotaxime; Ery: erythromycin; NT: non-typeable; Pen: penicillin; ST: sequence type.
This study, conducted after the introduction of systematic vaccination with PCV13 in the Andalusian child immunisation schedule, constitutes the first national publication on the molecular epidemiology of pneumococcal nasopharyngeal colonisation in healthy children with full PCV13 vaccination coverage. Several studies of pneumococcal carriage in children from various autonomous communities in Spain have been previously published, but these were conducted in settings with private vaccination and partial coverage, and the distribution of genotypes was only evaluated in two studies from the PCV7 period.8,9,12–15
PCVs have a profound impact on the pneumococcal population of the nasopharynx. They significantly reduce the circulation of VS, but fall short of completely eliminating them. This reduction is offset by an increase in the prevalence of NVS. As a result, pneumococcal colonisation rates similar16,17 or lower18,19 than those of the pre-vaccination era have been described after the introduction of PCVs, depending on whether the degree of replacement of VS by NVS was total or partial, respectively. We observed lower colonisation rates in the PCV13 period than those registered during the PCV7 period (19% vs 35%). This difference could be related to the unbalanced distribution between both study populations of some of the demographic characteristics, such as concomitant upper respiratory tract infection and recruitment in the first or third four-month period (less warm months), which were independent predictors of pneumococcal colonisation by multivariate analysis.
As expected, VS colonisation decreased very markedly, by 79%, between the PCV13 and PCV7 periods. Seven of the 10 circulating VS were eradicated during the historical period, including serotype 19A identified in 10% of the isolates from this last period, while only significant persistence (8%) of serotype 19F was observed. On the other hand, serotypes 3 and 6B, the proportions of which decreased between the two time periods, were detected very infrequently (1%) in the PCV13 period, although this finding was not statistically significant. This is not unexpected, because serotype 19F is one of the most frequently detected VS in colonisation studies performed after the introduction of PCV13.12,16,20 Serotype 19F was genetically diverse and associated with multiple antibiotic resistance. This serotype has a great capacity to form a biofilm and, therefore, a high capacity to evade the immune system, which, together with antibiotic resistance, are factors that may have contributed to its persistent circulation.21,22
There was a wide distribution of NVS in the PCV13 period, with 24 different serotypes identified. The dominant NVS included serotypes 15B/C, 11A, 23A, 10A, 16F, 31, NT, 23B and 21, which are common NVS in colonisation studies of the PCV13 period, although with some differences in the prevalence ranking between the various geographical areas.12,16,20,23,24 These serotypes have been associated with low invasive potential (OR < 1 in the comparison of the prevalence in IPD and carriers) in the PCV13 period, except for serotype 10A, which had an OR in the range of 1.2–1.7 in some studies.17,19,25 The 20-valent PCV, approved for the prevention of pneumonia and IPD in adult patients and likely to be marketed soon in the paediatric population, includes in its formulation this latter serotype and serotypes 11A and 15B, of high prevalence in this study.
The proportions of serotypes 11A and 31 increased significantly between the PCV13 and PCV7 periods due to the expansion of clones (ST6521 and ST1766, respectively) not detected in the PCV7 period study. ST6521 arose by recombination between clones 11A-ST62 and NT-ST344 and the global clone Spain9V-ST156, associated with serotypes 9V and 14.26 The clonal expansion of ST6521 is of concern due to its phenotypic characteristics of resistance to ampicillin/amoxicillin, greater ability to produce biofilm and increased ability to evade activation of the classical complement pathway and phagocytosis compared to ST838, which is also from Spain9V- ST156.27 In a context of antibiotic pressure and vaccine immunity, these properties may have contributed to its increasing relevance in the aetiology of acute otitis media in children and acute exacerbations of COPD in adults in certain geographical locations in Spain.28,29 In addition, this serotype can be associated with IPD in children and adults due to its high prevalence in carriers and its marked lethality: it is even more lethal than serotype 3.30,31
The rates of reduced susceptibility to penicillin and cefotaxime remained stable and a significant decrease in the rates of resistance to erythromycin was observed between the PCV13 and PCV7 periods. Although several VS (6A, 9V, 19A and 23F) associated to a variable degree with decreased susceptibility to β-lactam antibiotics were eradicated in the PCV7 period, this beneficial effect was offset by the appearance and clonal expansion of ST11A-ST6511.This is a pattern of evolution of antimicrobial resistance described after the introduction of PCVs, such that the decrease in resistance rates derived from the eradication of VS tends to be offset over time by the progressive acquisition of resistance determinants in NVS.32,33 It should also be noted that the SARS-CoV-2 pandemic has caused the percentage of resistant strains of pneumococcus to increase. This is clearly seen in serotype 11A, which has gone from having a MIC90 to penicillin of 2 μg/ml in Spain in the pre-pandemic period (2016–2019) to a MIC90 to penicillin of 4 μg/ml in 2020, which is a value considered to be resistant.34
This study has several limitations. First, as already mentioned, the different demographic characteristics of the study populations from the PCV7 and PCV13 periods are a confounding factor for the comparative analysis of colonisation rates. Second, the distribution of some serotypes may differ between healthy children and those with upper respiratory infection.35 Lastly, multiple colonisations were not evaluated by molecular techniques, nor was pre-enrichment performed on the culture, which, if it had been, could have increased the detection capacity of pneumococci in nasopharyngeal colonisation.
In conclusion, in the years after the introduction of PCV13 in the Andalusian paediatric immunisation schedule, a very residual circulation of VS was observed in nasopharyngeal colonisers, except for serotype 19F, as well as clonal expansion among NVS from ST11A-ST6511, with pathogenic capacity and associated with decreased susceptibility to penicillin and resistance to ampicillin. It would be advisable to maintain epidemiological surveillance of pneumococcal colonisation to monitor the evolution of NVS and antibiotic resistance rates.
FundingThis study was funded by a research project grant from Pfizer to IO (grant 5323485). The sponsor did not participate in any way in the design of the study, the collection, analysis and interpretation of the data, the writing of the article or in the decision to submit the article for publication.
Conflicts of interestIO received financial support from Pfizer to register for conferences and to conduct this project. ISC has received grants or fees as consultant/advisor and speaker, as well as financial support for attending conferences and practical courses from Pfizer, GlaxoSmithKline, Sanofi Pasteur, MSD and AstraZeneca. CC has received financial support for attending conferences from Pfizer, GlaxoSmithKline and MSD. The other authors have no conflicts of interest.
The authors wish to express their gratitude to Ignacio Cruz Navarro, Irene Bullón Durán and Esther Díaz Carrión for their collaboration in collecting samples from study participants.