Early detection of patients carrying multiresistant bacteria is an effective implement in surveillance programs. Our objective was to compare the semi-automatic Uroquattro HB&L “ESBL/AmpC Screening” (Alifax®) system with the routine culture on selective media to detect ESBL/pAmpC-producing microorganisms (3CGRE).
MethodsA total of 201 rectal swabs samples were processed by inoculating them into the Uroquattro HB&L system, performing growth curve measurements at 6.5 and 10h, and into direct culture medium.
ResultsThirty-five samples yielded 3CGRE. Measurements at 10h incremented the positive 3GCRE detection 5.7% in comparison with routine culture medium. In negative rectal swabs, the overall percent agreement at 6.5h and 10h versus routine culture medium was 93% and 90%, respectively.
ConclusionsThe Uroquattro HB&L system increased the detection of ESBL/pAmpC-producing bacteria compared to direct plating with an incubation time of 10h and shortens the time to report a negative sample.
La detección temprana de pacientes portadores de bacterias multirresistentes es una medida eficaz de los programas de vigilancia. Nuestro objetivo fue comparar el sistema semiautomático Uroquattro HB&L™ «ESBL/AmpCscreening» (Alifax®) frente al cultivo habitual en medios selectivos para detectar microorganismos productores de beta-lactamasas de espectro extendido (BLEE)/AmpC (3CGRE).
MétodosSe procesaron 201 frotis rectales mediante inoculación en el sistema Uroquattro HB&L™, se midió el crecimiento a las 6,5 y 10h, y en el medio de cultivo directo.
ResultadosTreinta y cinco muestras fueron positivas para 3CGRE. La lectura a las 10h incrementó la detección un 5,7% en comparación con el medio habitual. En muestras rectales negativas, la concordancia de la lectura global a las 6,5 y 10h con el medio de cultivo habitual fue del 93 y 90%, respectivamente.
ConclusionesEl sistema Uroquattro HB&L™ incrementó la detección de bacterias productoras de BLEE/pAmpC en comparación con el cultivo directo con un tiempo de incubación de 10h y acorta los tiempos de detección de muestras negativas.
Enterobacterales resistant to third-generation cephalosporins due to the acquisition of extended-spectrum beta-lactamases and plasmid-mediated cephalosporinases (ESBLs/pAmpC) currently constitute one of the most pressing medical problems worldwide. The growing dissemination of carbapenemase producers, many of them also resistant to third-generation cephalosporins, and the co-resistance pattern of these microorganisms represents an added problem.1 Early detection of patients carrying ESBL-/pAmpC-/carbapenemase-producing Enterobacterales (3GCRE) using surveillance cultures on admission and isolation of colonized patients is a major strategy designed to prevent the spread of resistance genes.2
Recent advances in the detection of 3GCRE are based on molecular techniques or chromogenic selective media.3 Molecular techniques continue to be the gold standard due to their high, almost 100% sensitivity and specificity. All of them are based on amplification of the main genes involved in these enzymes in 1–3h.4 Nevertheless, the cost of molecular methods and the limited targets available restrict their systematic implementation. Chromogenic selective media are the cheapest and easiest technique for detection of 3GCRE and more cost effective5 despite their lower sensitivity (around 80%) and the longer time taken to obtain results (between 24 and 48h).5
Another strategy that improves detection of colonized patients is to add a pre-enrichment step.6 The main inconvenience of enrichment is the delay associated with obtaining results.7 The Uroquattro HB&L (Alifax®) is a new system based on a selective pre-enrichment medium and continuous measurement of growth curves using light scattering technology, designed to increase the detection of third-generation cephalosporin-resistant Enterobacterales. The aim of this study was to compare a selective medium routinely used in our laboratory with the semi-automated Uroquattro HB&L system (Alifax®).
Material and methodsPatients and microbiological analysisThe study was performed at the University Hospital Virgen Macarena (a 900-bed tertiary hospital) in Seville, Spain. From June to October 2017, consecutive double rectal swabs were collected (Copan®, Italy) from 201 inpatients included in the local surveillance programme (upon ICU admission, previous colonization status, transfers from other centres and roommates of positive cases). One swab was plated on the routinely used medium (MacConkey agar supplemented with cefotaxime 4μg/ml) and incubated at 35°C for 18h. Results from routine medium were reported in 18–24h (for negative results when no growth of Enterobacterales was observed) and in 48h (for negative results with growth of non 3CGRE or positive results). The second swab was inoculated into the Uroquattro HB&L system (UHBL). UHBL is a semi-automatic analyser that uses a light scattering technology to test bacteria susceptibility on rectal swabs. UHBL measures the growth of bacteria in specific selective enrichment broths providing real time growth curves and bacterial counts (CFU/ml) every hour with a specific interpretation algorithm. Measurements of the growth curve were taken at 6.5h (according to the manufacturer's instructions) and at 10h (to assess whether longer incubation might increase detection). Positive samples were subcultured in MacConkey agar containing cefotaxime (4mg/l). All isolates detected by both methods were identified by MALDI-TOF (Bruker). Screening for ESBL or pAmpC production was performed on Mueller-Hinton agar plates with or without cloxacillin (200mg/l), with double discs according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines.
The presence of bla genes (ESBL: blaSHV, blaCTX-M-1, blaCTX-M-9; pAmpC: blaMOX, blaCIT, blaDHA, blaACC, blaEBC, blaCMY and blaFOX; carbapenemase: blaKPC, blaNDM, blaVIM, blaIMP and blaOXA-48) was studied by PCR using specific group primers4 and further sequencing of amplicons. The reference value was the combination of obtained from both methods.
Statistical analysisTo assess of concordant and discordant results, percent agreement and the exact McNemar test were calculated (SPSS Statistics, version 23.0) for 3GCRE positives and negatives rectal swab of UHBL system at 6.5 and at 10h and the routine medium.
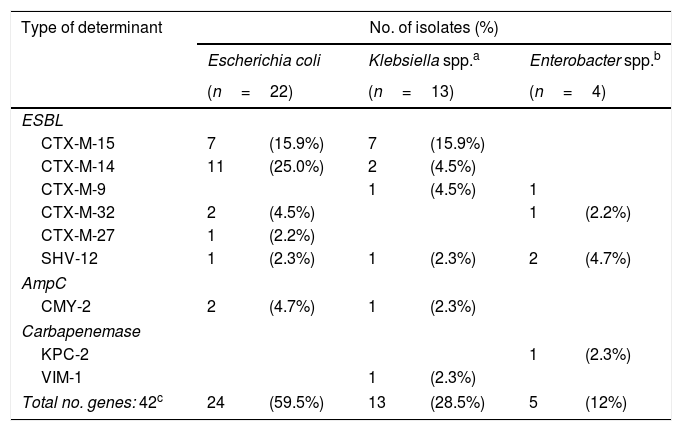
ResultsA total of 35 samples (17.4%) were positive for 3GCRE. E. coli (n=22) was the most prevalent microorganism (Table 1). Among ESBL enzymes, the most frequent was CTX-M-15 (n=14, 37.8%); pAmpC (CMY-2) and carbapenemases (KPC-2 and VIM-1) were also found (Table 1).
Distribution of Enterobacterales and enzymes among the 35 positive rectal swabs.
| Type of determinant | No. of isolates (%) | |||||
|---|---|---|---|---|---|---|
| Escherichia coli | Klebsiella spp.a | Enterobacter spp.b | ||||
| (n=22) | (n=13) | (n=4) | ||||
| ESBL | ||||||
| CTX-M-15 | 7 | (15.9%) | 7 | (15.9%) | ||
| CTX-M-14 | 11 | (25.0%) | 2 | (4.5%) | ||
| CTX-M-9 | 1 | (4.5%) | 1 | |||
| CTX-M-32 | 2 | (4.5%) | 1 | (2.2%) | ||
| CTX-M-27 | 1 | (2.2%) | ||||
| SHV-12 | 1 | (2.3%) | 1 | (2.3%) | 2 | (4.7%) |
| AmpC | ||||||
| CMY-2 | 2 | (4.7%) | 1 | (2.3%) | ||
| Carbapenemase | ||||||
| KPC-2 | 1 | (2.3%) | ||||
| VIM-1 | 1 | (2.3%) | ||||
| Total no. genes: 42c | 24 | (59.5%) | 13 | (28.5%) | 5 | (12%) |
1 rectal swab=E. coli (CTX-M-15+CMY-2).
1 rectal swab=E. coli (CTX-M-14)+K. oxytoca (CTX-M-14).
1 rectal swab=E. gergoviae (KPC-2+SHV-12).
1 rectal swab=E. coli (CTX-M-15)+K. pneumoniae (CTX-M-14).
2 rectal swabs=E. coli (CTX-M-14)+K. pneumoniae (CTX-M-15).
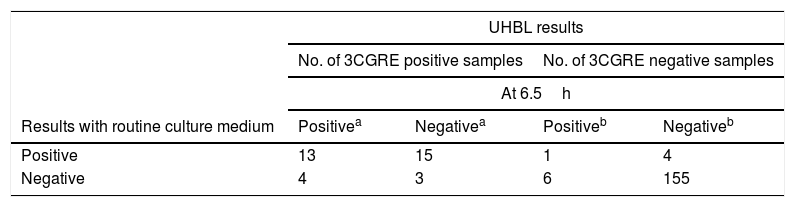
Measurements of the UHBL system at 6.5h and 10h were compared to direct culture on solid medium for all 3GCRE positive and negative cultures (Table 2). Thirty-five rectal swabs were positive for 3GCRE for both UHBL system and routine medium, whereas 4 and 7 rectal swabs were positive only with UHBL system at 6.5h and 10h, respectively (Table 2). Measurements at 10h incremented a 5.7% (95% confidence interval [CI], 1.5–1.8%) (2/35) the positive 3GCRE detection in comparison with the routine culture medium. The increments of false positive results were 1.2% (95% confidence interval [CI], 0.3–0.4%) (2/166), and 7.2% (95% confidence interval [CI], 4.1–12.2%) (12/166), at 6.5h and 10h, respectively.
Comparison of the UHBL* system at 6.5h and 10h with the routine culture medium.
| UHBL results | ||||
|---|---|---|---|---|
| No. of 3CGRE positive samples | No. of 3CGRE negative samples | |||
| At 6.5h | ||||
| Results with routine culture medium | Positivea | Negativea | Positiveb | Negativeb |
| Positive | 13 | 15 | 1 | 4 |
| Negative | 4 | 3 | 6 | 155 |
| At 10h | ||||
|---|---|---|---|---|
| Results with routine culture medium | Positivec | Negativec | Positived | Negatived |
| Positive | 23 | 5 | 3 | 2 |
| Negative | 7 | 0 | 14 | 147 |
Positive results were considered the detection of ESBL/pAmpC producers. Negative results were considered lack of growth or growth of third-generation cephalosporins resistant Gram-negative but not ESBL/pAmpC producers.
Overall percent agreement, 45.7% (95% confidence interval [CI], 30.4–61.8%); exact McNemar test, p=0.74.
Overall percent agreement, 93% (95% confidence interval [CI], 89.2–96.7%); exact McNemar test, p=0.01.
Overall percent agreement, 65% (95% confidence interval [CI], 49.1–79.1%); exact McNemar test, p=0.77.
Active screening cultures to prevent nosocomial infections and outbreaks are performed in many hospitals,8 although an optimal screening protocol has not yet been established.8 The present study evaluates for the first time a semi-automated screening system, the Uroquattro HB&L, which provides early and higher detection of faecal carriers of ESBL-/pAmpC-/carbapenemase-producing Enterobacterales.
There are two aspects of carrier detection surveillance that are fundamental: firstly, the accuracy of the method and secondly, the time to detection.9 The use of a pre-enrichment step increases the detection efficiency in comparison to the usual strategy of inoculating samples directly onto selective solid media, at the expenses of longer detection time and higher false positive rates. The results obtained in a previous study comparing solid and pre-enrichment cultures10 found an agreement in negative samples of 96.8%, consistent with our results. Previous studies yielded higher increases of detection with the enrichment (25.9%10 and 20%11 compared with 5.7% in our study) and lower increases of false positive rates (2.1%10 and 0.5%11 compared with our study), taken into account lower prevalence of ESBL producer positive patients (4.8%10 and 9%,11 compared to 18.4% in our study). The variation between the yield and the false positive rate with pre-enrichment, may increase when the prevalence of ESBL/AmpC carriers rise. Rectal swabs negative by direct culture and positive by enrichment may be due to low bacterial loads, which may have less epidemiological relevance, although these bacterial loads may variate depending on time of hospitalization and antibiotic exposure. Selection of previous detected ESBL producers have been seen in patients with negative samples without enrichment upon admission of the ICU.12 Follow-up studies are necessary to establish the risk of transmission according to faecal load.
Uroquattro HB&L system reduces the time of detection to 6.5h, according to the manufacturer, although an increment in detection was obtained at 10h according to our results. Nevertheless, both 6.5h and 10h enable the characterization of 3CGRE to begin on the same day that the sample is received and, a more important issue, shortening the time for negative results. Earlier detection could have an impact on costs. Sypsa et al.,13 using a mathematical model, concluded that a delay in detection, even by one day, increased the costs associated with outbreaks. On the other hand, an earlier negative result may have an impact by reducing the adverse effects of isolation, such as supportive care failures due less time spent by health workers or patient mental problems.14 Molecular methods could reduce even more the result time, but a higher cost.15
This study had limitations. In our study we used the Copan Transystem™ swabs with Amies solid medium, while the manufacturer of the semi-automated system recommends the use of nylon-flocked swabs with Amies liquid (ESwab). However, not all hospitals have introduced the use of the ES-swab®, and our results could be valid in these situations. Another limitation is that we did not inoculated the rectal swabs on blood agar as a control of enteric microbiota. In addition, this system does not detect OXA-48 producers not co-producing ESBL/AmpC, but the prevalence of infections due to these microorganisms is very low in our area.
In conclusion, the use of a preenrichment semi-automatic system Uroquattro HB&L in rectal swabs improved the detection of ESBL/pAmpC faecal carriers with an incubation time of 10h, shortening the reporting time of negative results.
FundingThis work was partially supported by Plan Nacional de I+D+i 2013–2016 and Instituto de Salud Carlos III, subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía, Industria y Competitividad, Spanish Network for Research in Infectious Diseases (RD16/0016/0001) co-financed by European Development Regional Fund “A way to achieve Europe”, Operative program Intelligent Growth 2014–2020.
Conflict of interestsThe authors declare no conflict of interest.