The rapid identification of bacteraemia-causing pathogens could assist clinicians in the timely prescription of targeted therapy, thereby reducing the morbidity and mortality of this infection. In recent years, numerous techniques that rapidly and directly identify positive blood cultures have been marketed, with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) being one of the most commonly used.
MethodsThe aim of this systematic review and meta-analysis was to evaluate the accuracy of MALDI-TOF (Bruker®) for the direct identification of positive blood culture bottles.
ResultsA meta-analysis was performed to summarize the results of the 32 studies evaluated. The overall quality of the studies was moderate. For Gram-positive bacteria, overall rates of correct identification of the species ranged from 0.17 to 0.98, with a cumulative rate (random-effects model) of 0.72 (95% CI: 0.64–0.80). For Gram-negative bacteria, correct identification rates ranged from 0.66 to 1.00, with a cumulative effect of 0.92 (95% CI: 0.88–0.95). For Enterobacteriaceae, the rate was 0.96 (95% CI: 0.94–0.97).
ConclusionMALDI-TOF mass spectrometry shows high accuracy for the correct identification of Gram-negative bacteria, particularly Enterobacteriaceae, directly from positive blood culture bottles, and moderate accuracy for the identification of Gram-positive bacteria (low for some species).
La identificación rápida de los patógenos causantes de bacteriemia orienta a los clínicos a prescribir con mayor celeridad un tratamiento dirigido y reducir así la morbimortalidad de dicha infección. Durante los últimos años, han aparecido en el mercado numerosas técnicas con la intención de cubrir esta necesidad, que logran una identificación rápida y directa a partir de los frascos de hemocultivos positivos. La espectrometría de masas matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry (MALDI-TOF MS) es una de las tecnologías más utilizadas en este campo.
MétodosEl objetivo de ese estudio es realizar una revisión sistemática y metaanálisis que evalúe la precisión de MALDI-TOF MS (Bruker) para la identificación directa a partir de frascos de hemocultivos positivos.
ResultadosEl metaanálisis fue realizado para sintetizar los resultados de los 32 estudios evaluados. La calidad total de los estudios fue moderada. Para las bacterias grampositivas, el ratio total de identificaciones correctas a nivel de especie fue del 0,17 al 0,98 con un ratio acumulativo (modelo de efectos aleatorios) de 0,72 (IC 95%: 0,64-0,80). Para las bacterias gramnegativas, el rango de identificaciones correctas fue del 0,66 al 1,00 con un efecto acumulativo de 0,92 (IC 95%: 0,88-0,95), llegando a un 0,96 (IC 95%: 0,94-0,97) en Enterobacteriaceae.
ConclusionesLa espectrometría de masas MALDI-TOF muestra una alta precisión para la correcta identificación de bacterias gramnegativas realizada directamente a partir de los frascos de hemocultivos positivos, siendo mayor en el grupo de las enterobacterias. Para grampositivas, la precisión es moderada, llegando a ser baja para alguna especie.
Bloodstream infection (BSI) is one of the leading causes of death around the world, with an estimated incidence of up to 19 million people worldwide every year.1 Rapid identification of the causative organism is essential to guide clinicians in the selection of the most appropriate targeted treatment for patients with BSI, and it is associated with improved patient outcomes.2 Many remarkable improvements have been made in the attempt to reduce the time required to identify the pathogen in positive blood cultures, including the direct inoculation of fluids from positive blood cultures into automated systems, fluorescent in situ hybridization (FISH), and PCR. All these methods, however, are expensive and require several hours of work.3
In recent years, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has emerged as the new standard in bacterial identification and is now being adopted in clinical laboratories worldwide.
MALDI-TOF technology uses a laser to softly ionize the many structural elements (primarily ribosomal proteins) of bacteria and yeasts and then separate the molecules according to their mass-to-charge ratio (m/z).4,5 Four commercial systems are currently in use worldwide: the MALDI-TOF BioTyper (Bruker Daltonics, Bremen, Germany); Saramis (AnagnosTec, Potsdam, Germany); Andromas (Andromas, Paris, France), and Vitek MS (bioMérieux, Marcy l’Étoile, France), although Bruker's BioTyper system is the most widely used in clinical practice and research.
Different studies have reported successful identification of bacteria directly from positive blood cultures using MALDI-TOF MS. Nevertheless, the results of these studies have varied, depending on the distribution of microbial isolates, the pre-treatment/extraction method applied, and where the log score cut-off values were defined.6 One of these methods has become commercially available as the Sepsityper Kit (Bruker Daltonics) to standardize the preparation of blood culture prior to spectrometric analysis. The method involves lysis of blood cells, followed by centrifugation and washing steps. The final result is a pellet of bacteria, which is further processed by standard methods for identification using MALDI-TOF.7,8
One disadvantage of the Sepsityper kit is the additional cost, so that many laboratories have developed their own in-house methods of bacterial extraction from positive blood culture bottles.9–11 These methods vary in their approach to removing human cellular components and enriching the microbes from blood culture fluids. Saponin12 and ammonium chloride13 to lyse the blood cells have been described. Separation of microorganisms from blood cells can be performed with differential centrifugation and gel separator tubes.14,15 Simple stepwise sedimentation of blood cells and microorganisms has also been described.5,16,17
The present systematic review and meta-analysis aimed to evaluate the accuracy of MALDI-TOF for the identification of bacteria directly from positive blood culture bottles. We analyzed the different pre-treatment methods and other variables that may have influenced the correct identification of bacteria to species level from positive blood culture bottles.
MethodsWe designed a systematic review and meta-analysis of the scientific literature. Our study was performed according to the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines for systematic reviews.18
Information sources and searchesWe systematically searched the Medline (PubMed), Embase, Cochrane Database of Systematic Reviews (CDSR), Center for Reviews and Dissemination (CRD), Network of Agencies for Health Technology Assessment database, and the clinical trial results database, “ClinicalTrials.gov”, to identify studies between January 1, 2011 and October 1, 2015 that assessed the accuracy of MALDI-TOF MS for direct identification of bacteria from blood cultures. To localize other undetected published articles, a manual search was performed for references in relevant studies and specialized medical journals. No language restrictions were applied. To achieve maximum sensitivity, the search was performed combining the following keywords and free terms: “bacteraemia”, “sepsis”, “bloodstream infection”, “humans”, “matrix-assisted laser desorption ionization time-of-flight mass spectrometry”, “MALDI-TOF mass spectrometry”, “clinical trial”, “prospective studies”, “accuracy”, “sensitivity and specificity”, “comparative study”, “evaluation studies”, and “diagnosis”.
Eligibility criteria and study selectionTwo investigators screened the titles and abstracts of the references localized. The full text of potentially eligible studies was read and evaluated for definitive inclusion. Included were studies of MALDI-TOF MS (Bruker) performed to identify bacteria from blood cultures of patients, compared with routine bacterial identification (automated or manual methods: phenotypic, microbiological, molecular diagnosis). Only studies using the Bruker® system were included because it is the most frequently used system and allows for greater standardization in evaluation. Exclusion criteria were: studies that did not investigate blood cultures; studies identifying bacteria by mass spectrometry methods different from MALDI-TOF MS; studies without a comparator method, studies applying MALDI-TOF to subcultures of positive blood cultures; studies for the identification of mycobacteria, yeasts or parasites. We also excluded non-original articles, non-human studies, in vitro simulation studies, case-series studies, editorials and letters.
Data extraction and assessment of risk biasTwo independent researchers reviewed all references in order to identify articles that required full-text appraisal, with the final decision reached through consensus. All retrieved articles were evaluated for inclusion in the systematic review and meta-analysis. Study characteristics (design, year of publication, study country, period of study), population (size, age, sample type), intervention (previous processing, bacterial database version, score, procedures), comparator (gold standard, different comparators), and outcomes (proportion of identification, accuracy) were recorded.
We used the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) checklist to assess the quality of the studies and potential risk of bias.19 Two researchers independently performed the quality assessment.
Statistical analysisThe main effect size index was the correct identification rate (CIR); specifically, the identification rate obtained by use of MALDI-TOF was compared with results obtained from the reference method. Some researchers have found that using a lower threshold (cut-off 2.0) than the one specified by the manufacturer provided the best identification compared to the reference method.
Statistical significance was defined as a p value of ≤0.001. Meta-analyses were performed with R statistical software (version 3.0.2; R Foundation for Statistical Computing, Vienna, Austria) and the package “meta” (version 3.1-2). The Freeman-Tukey transformation of inverse hyperbolic sine function was used to calculate the CIR. Both fixed-effect and random-effects (DerSimonian and Laird method) meta-analyses were performed and heterogeneity was evaluated on the basis of I2, the heterogeneity measure of Cochran's Q test. Heterogeneity was evaluated quantitatively using I2 in which I2 values of 25%, 50%, and 75% indicate low, moderate, and high heterogeneity, respectively. When significant heterogeneity was observed among the included studies, the random-effects model was considered; otherwise, the fixed-effect model was used. In order to check whether publication bias might have influenced the validity of the results, funnel plots and Egger's test were applied.
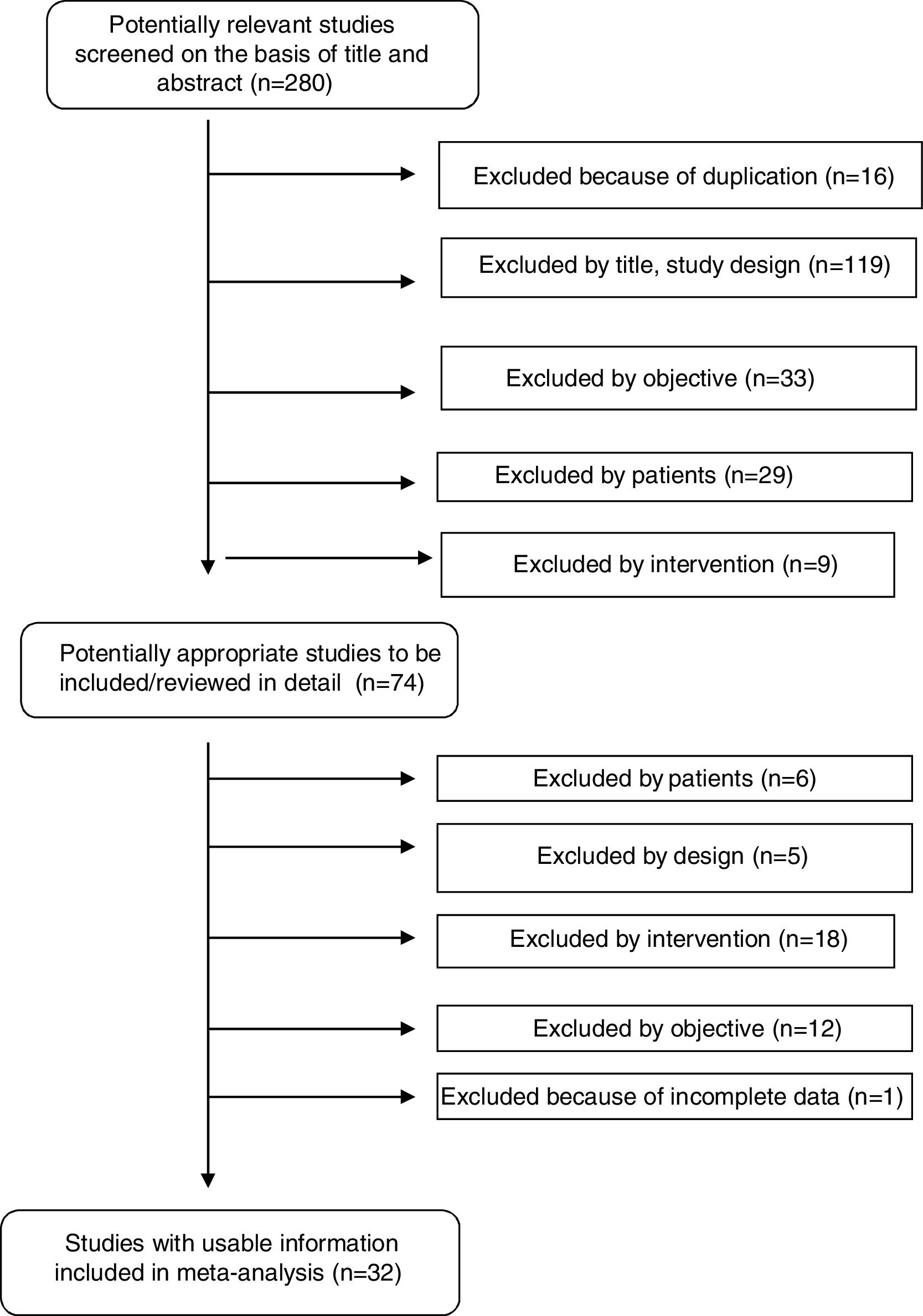
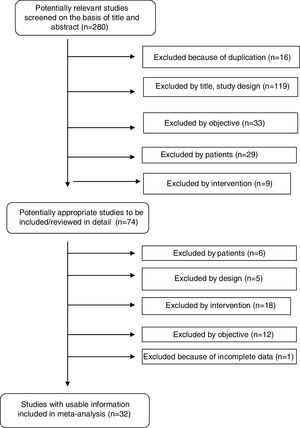
ResultsStudy selectionOur systematic review identified a total of 280 references. After duplicates (16) were ruled out and titles and abstracts (264) screened, 74 potentially relevant studies were retrieved for inclusion criteria and data extraction. Forty-two studies were excluded for various reasons: data not shown, no intervention, use of a technique different from MALDI-TOF (Bruker), no proper comparator, and no clinical samples. A total of 32 studies fulfilled all inclusion criteria and were finally selected for performance of the systematic review and meta-analysis. Fig. 1 shows a flowchart of the process.
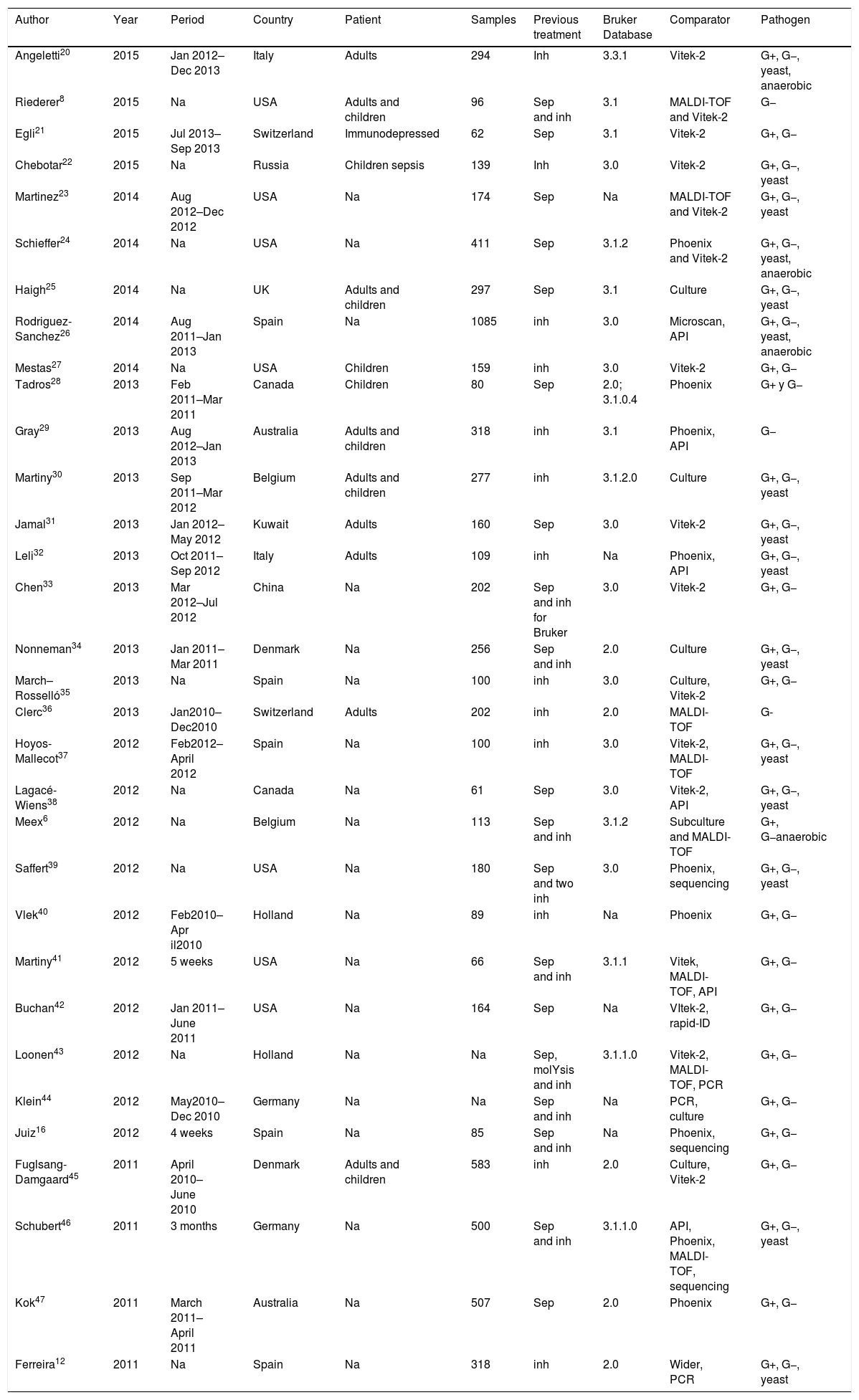
Study characteristicsThe main characteristics of the eligible studies are listed in Table 1. Thirty-two articles were finally included in this meta-analysis: 19 from Europe, 9 performed in North America (7 in USA and 2 in Canada), 2 carried out in Australia, and 2 in Asia. The population consisted of adults in four studies and children in 3 studies; in 19 studies, the population was not specified.
Main characteristics of 32 studies included in the systematic review and meta-analysis.
| Author | Year | Period | Country | Patient | Samples | Previous treatment | Bruker Database | Comparator | Pathogen |
|---|---|---|---|---|---|---|---|---|---|
| Angeletti20 | 2015 | Jan 2012–Dec 2013 | Italy | Adults | 294 | Inh | 3.3.1 | Vitek-2 | G+, G−, yeast, anaerobic |
| Riederer8 | 2015 | Na | USA | Adults and children | 96 | Sep and inh | 3.1 | MALDI-TOF and Vitek-2 | G− |
| Egli21 | 2015 | Jul 2013–Sep 2013 | Switzerland | Immunodepressed | 62 | Sep | 3.1 | Vitek-2 | G+, G− |
| Chebotar22 | 2015 | Na | Russia | Children sepsis | 139 | Inh | 3.0 | Vitek-2 | G+, G−, yeast |
| Martinez23 | 2014 | Aug 2012–Dec 2012 | USA | Na | 174 | Sep | Na | MALDI-TOF and Vitek-2 | G+, G−, yeast |
| Schieffer24 | 2014 | Na | USA | Na | 411 | Sep | 3.1.2 | Phoenix and Vitek-2 | G+, G−, yeast, anaerobic |
| Haigh25 | 2014 | Na | UK | Adults and children | 297 | Sep | 3.1 | Culture | G+, G−, yeast |
| Rodriguez-Sanchez26 | 2014 | Aug 2011–Jan 2013 | Spain | Na | 1085 | inh | 3.0 | Microscan, API | G+, G−, yeast, anaerobic |
| Mestas27 | 2014 | Na | USA | Children | 159 | inh | 3.0 | Vitek-2 | G+, G− |
| Tadros28 | 2013 | Feb 2011–Mar 2011 | Canada | Children | 80 | Sep | 2.0; 3.1.0.4 | Phoenix | G+ y G− |
| Gray29 | 2013 | Aug 2012–Jan 2013 | Australia | Adults and children | 318 | inh | 3.1 | Phoenix, API | G− |
| Martiny30 | 2013 | Sep 2011–Mar 2012 | Belgium | Adults and children | 277 | inh | 3.1.2.0 | Culture | G+, G−, yeast |
| Jamal31 | 2013 | Jan 2012–May 2012 | Kuwait | Adults | 160 | Sep | 3.0 | Vitek-2 | G+, G−, yeast |
| Leli32 | 2013 | Oct 2011–Sep 2012 | Italy | Adults | 109 | inh | Na | Phoenix, API | G+, G−, yeast |
| Chen33 | 2013 | Mar 2012–Jul 2012 | China | Na | 202 | Sep and inh for Bruker | 3.0 | Vitek-2 | G+, G− |
| Nonneman34 | 2013 | Jan 2011–Mar 2011 | Denmark | Na | 256 | Sep and inh | 2.0 | Culture | G+, G−, yeast |
| March–Rosselló35 | 2013 | Na | Spain | Na | 100 | inh | 3.0 | Culture, Vitek-2 | G+, G− |
| Clerc36 | 2013 | Jan2010–Dec2010 | Switzerland | Adults | 202 | inh | 2.0 | MALDI-TOF | G- |
| Hoyos-Mallecot37 | 2012 | Feb2012–April 2012 | Spain | Na | 100 | inh | 3.0 | Vitek-2, MALDI-TOF | G+, G−, yeast |
| Lagacé-Wiens38 | 2012 | Na | Canada | Na | 61 | Sep | 3.0 | Vitek-2, API | G+, G−, yeast |
| Meex6 | 2012 | Na | Belgium | Na | 113 | Sep and inh | 3.1.2 | Subculture and MALDI-TOF | G+, G−anaerobic |
| Saffert39 | 2012 | Na | USA | Na | 180 | Sep and two inh | 3.0 | Phoenix, sequencing | G+, G−, yeast |
| Vlek40 | 2012 | Feb2010–Apr il2010 | Holland | Na | 89 | inh | Na | Phoenix | G+, G− |
| Martiny41 | 2012 | 5 weeks | USA | Na | 66 | Sep and inh | 3.1.1 | Vitek, MALDI-TOF, API | G+, G− |
| Buchan42 | 2012 | Jan 2011–June 2011 | USA | Na | 164 | Sep | Na | VItek-2, rapid-ID | G+, G− |
| Loonen43 | 2012 | Na | Holland | Na | Na | Sep, molYsis and inh | 3.1.1.0 | Vitek-2, MALDI-TOF, PCR | G+, G− |
| Klein44 | 2012 | May2010–Dec 2010 | Germany | Na | Na | Sep and inh | Na | PCR, culture | G+, G− |
| Juiz16 | 2012 | 4 weeks | Spain | Na | 85 | Sep and inh | Na | Phoenix, sequencing | G+, G− |
| Fuglsang-Damgaard45 | 2011 | April 2010–June 2010 | Denmark | Adults and children | 583 | inh | 2.0 | Culture, Vitek-2 | G+, G− |
| Schubert46 | 2011 | 3 months | Germany | Na | 500 | Sep and inh | 3.1.1.0 | API, Phoenix, MALDI-TOF, sequencing | G+, G−, yeast |
| Kok47 | 2011 | March 2011–April 2011 | Australia | Na | 507 | Sep | 2.0 | Phoenix | G+, G− |
| Ferreira12 | 2011 | Na | Spain | Na | 318 | inh | 2.0 | Wider, PCR | G+, G−, yeast |
Sep, Sepsityper; inh, in house; Na, not available; G+, Gram-positive; G-, Gram-negative.
The global number of analyzed samples was 7187. In two studies,43,44 the number of samples was not provided. All studies used clinical isolates. All included studies reported their pre-treatment method: 13 used an in-house method,12,20,22,26,27,29,30,32,35–37,40,45 7 used Sepsityper,21,24,25,28,31,38,42 and the rest used a combination of the two.6,8,16,33,34,39,41,43,44,46
Twenty-five of the 32 studies included used a commercial identification system as the comparator method and in the other 7 studies,6,25,30,34,35,44,45 the comparator was identification by MALDI-TOF from subcultures.
Risk of bias of studies includedThe overall quality of the studies included was moderate (Supplementary Table S1). All but two studies26,29 recorded the selection criteria for patients. Only one study reported the simultaneous realization of both tests.24 An adequate gold standard was used as comparator in all studies. Blinding of investigators for the interpretation of the reference test was reflected in only two studies.24,34 Losses that took place during the studies were explained and uninterpretable results were reported in all studies.
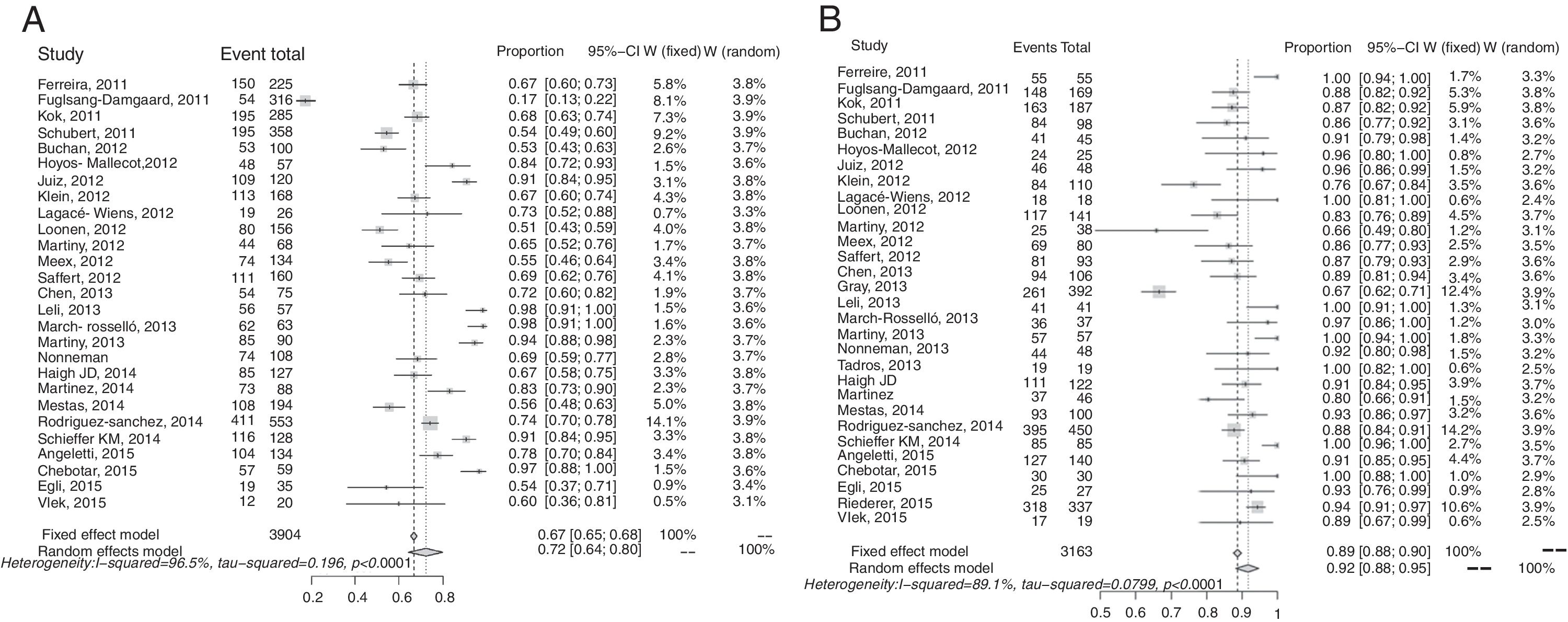
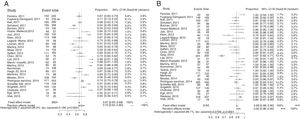
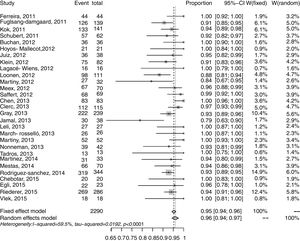
Synthesis of resultsOverall results for Gram-negative bacteria were reported in 30 studies, and 3163 recorded samples. The correct identification rate (CIR) for these pathogens in the various studies ranged from 0.66 to 1.00 at the species level. There was significant heterogeneity in this meta-analysis (I2: 89.1%; p<0.001) (Fig. 2). The pooled CIR estimated using a random-effects model was 0.92 (95% CI: 0.88–0.95). Publication bias in this meta-analysis was not significant (p=0.02 by Egger's test; Supplementary Fig. S2).
The forest plot for the meta-analysis: overall identification of Gram-positive bacteria at the species level (A) and Gram-negative bacteria at the species level (B). CI, confidence interval; W, weight; fixed, fixed-effect model; random, random-effects model; events, number of correct identifications; total, total number of identifications. Gray squares represent the weight of individual studies with the fixed-effect model; horizontal lines through the squares represent 95% confidence intervals; gray diamonds represent the overall estimate and its confidence interval; dotted vertical lines represent the fixed-effect model; and dashed vertical lines represent the random-effects model.
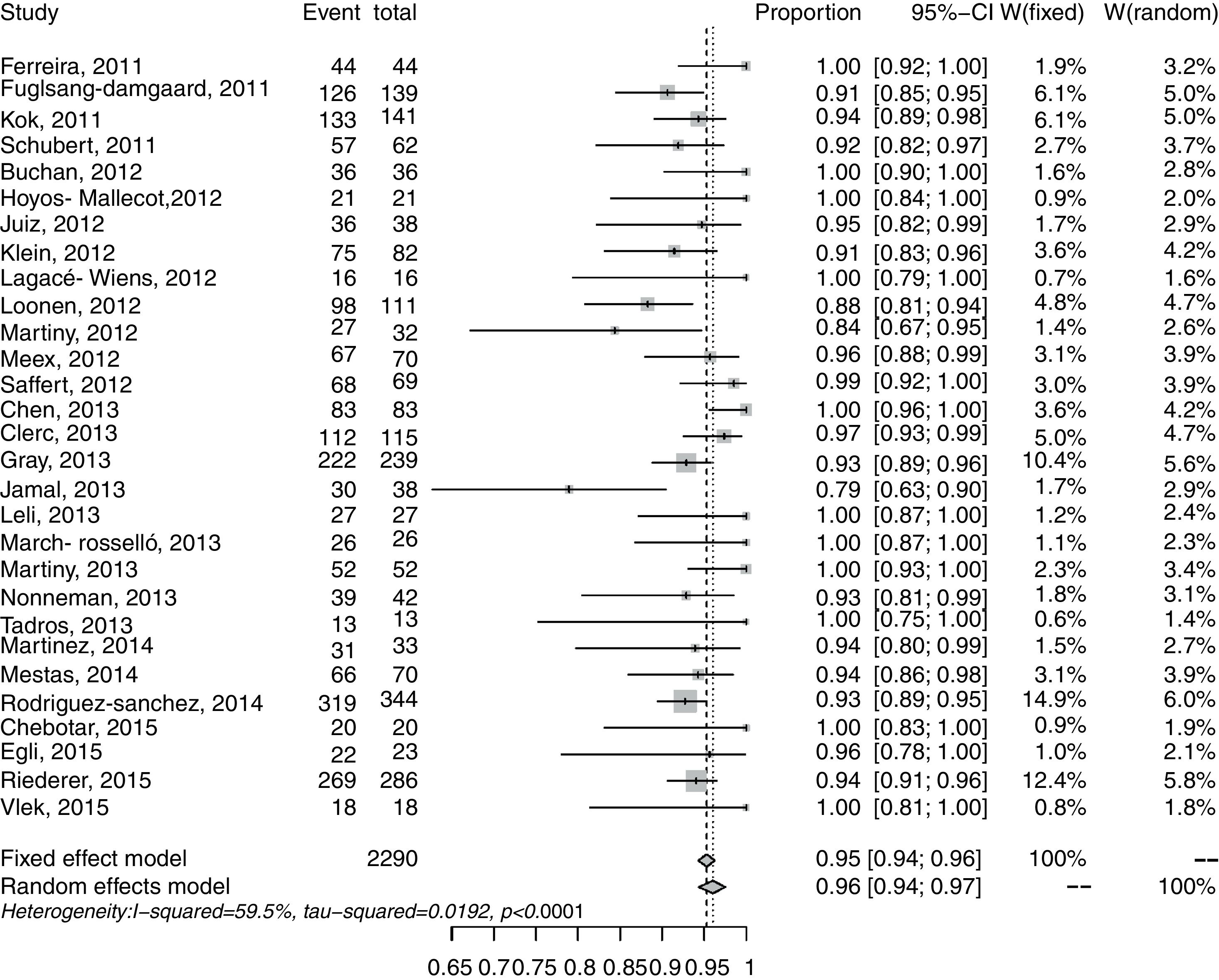
The results for correct identification of Enterobacteriaceae were recorded in 28 studies and 2293 samples. The CIRs in the different studies ranged from 0.79 to 1.00 at the species level. Heterogeneity was I2=59.5% (p<0.001; Fig. 2). The pooled CIR estimated with the random-effects model was 0.96 (95% CI: 0.94–0.97). Publication bias in this meta-analysis was not significant (p=0.06 by Egger's test; Supplementary Fig. S2).
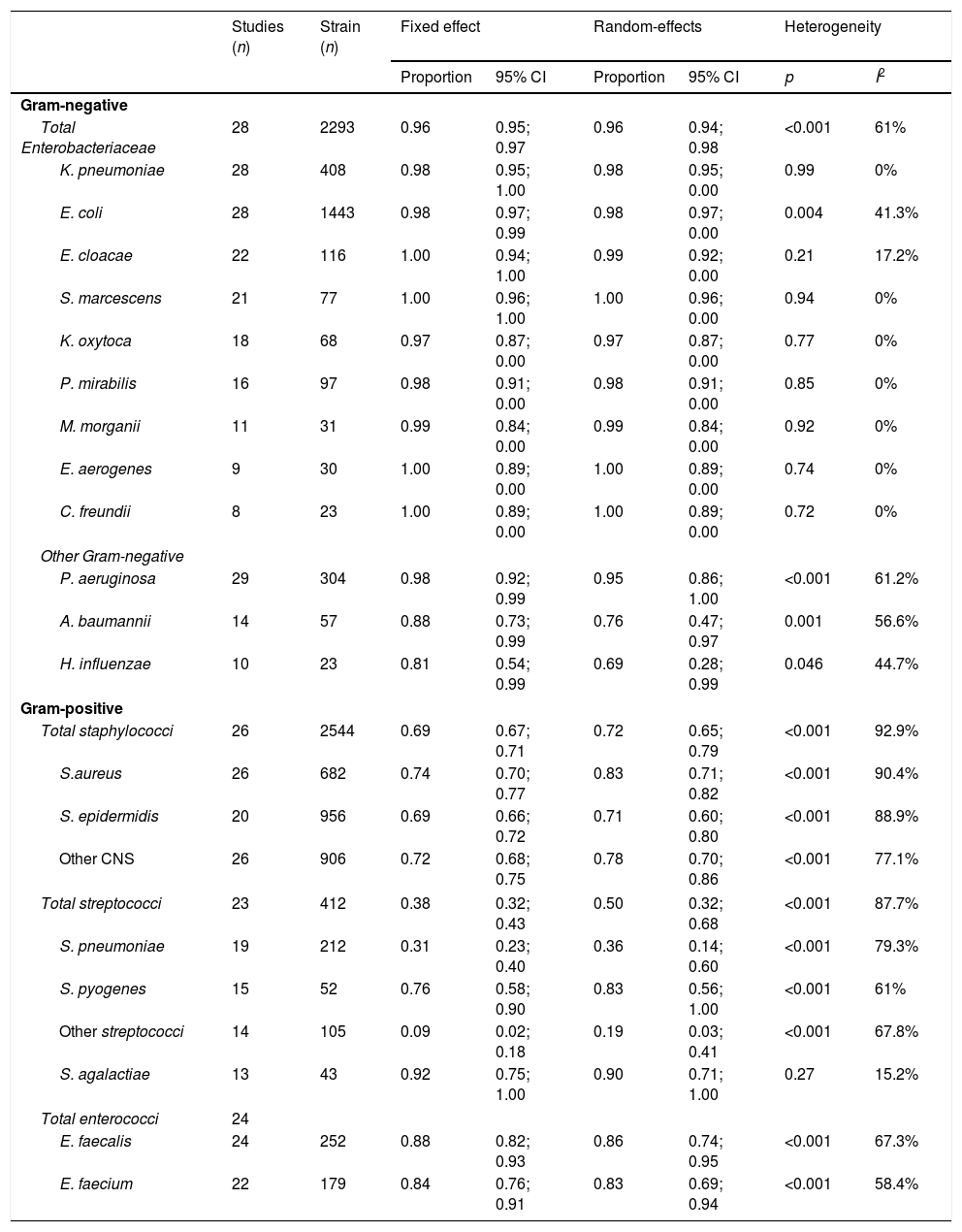
Accuracy of identification was also analyzed for each species of Enterobacteriaceae; the results are set out in Table 2. The highest correct identification rates (1.00) were of Enterobacter cloacae, Serratia marcescens, Enterobacter aerogenes, and Citrobacter freundii. In this analysis, heterogeneity in the species of Enterobacteriaceae was not significant, and I2 values ranged from 0% (p>0.70) to 41.3% (p=0.004). The CIRs for other Gram-negative pathogens were: Pseudomonas aeruginosa, 0.98; Acinetobacter baumannii, 0.88; and Haemophilus influenzae 0.81.
Meta-analysis of the accuracy of MALDI-TOF for included Gram-negative and Gram positive pathogens.
| Studies (n) | Strain (n) | Fixed effect | Random-effects | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| Proportion | 95% CI | Proportion | 95% CI | p | I2 | |||
| Gram-negative | ||||||||
| Total Enterobacteriaceae | 28 | 2293 | 0.96 | 0.95; 0.97 | 0.96 | 0.94; 0.98 | <0.001 | 61% |
| K. pneumoniae | 28 | 408 | 0.98 | 0.95; 1.00 | 0.98 | 0.95; 0.00 | 0.99 | 0% |
| E. coli | 28 | 1443 | 0.98 | 0.97; 0.99 | 0.98 | 0.97; 0.00 | 0.004 | 41.3% |
| E. cloacae | 22 | 116 | 1.00 | 0.94; 1.00 | 0.99 | 0.92; 0.00 | 0.21 | 17.2% |
| S. marcescens | 21 | 77 | 1.00 | 0.96; 1.00 | 1.00 | 0.96; 0.00 | 0.94 | 0% |
| K. oxytoca | 18 | 68 | 0.97 | 0.87; 0.00 | 0.97 | 0.87; 0.00 | 0.77 | 0% |
| P. mirabilis | 16 | 97 | 0.98 | 0.91; 0.00 | 0.98 | 0.91; 0.00 | 0.85 | 0% |
| M. morganii | 11 | 31 | 0.99 | 0.84; 0.00 | 0.99 | 0.84; 0.00 | 0.92 | 0% |
| E. aerogenes | 9 | 30 | 1.00 | 0.89; 0.00 | 1.00 | 0.89; 0.00 | 0.74 | 0% |
| C. freundii | 8 | 23 | 1.00 | 0.89; 0.00 | 1.00 | 0.89; 0.00 | 0.72 | 0% |
| Other Gram-negative | ||||||||
| P. aeruginosa | 29 | 304 | 0.98 | 0.92; 0.99 | 0.95 | 0.86; 1.00 | <0.001 | 61.2% |
| A. baumannii | 14 | 57 | 0.88 | 0.73; 0.99 | 0.76 | 0.47; 0.97 | 0.001 | 56.6% |
| H. influenzae | 10 | 23 | 0.81 | 0.54; 0.99 | 0.69 | 0.28; 0.99 | 0.046 | 44.7% |
| Gram-positive | ||||||||
| Total staphylococci | 26 | 2544 | 0.69 | 0.67; 0.71 | 0.72 | 0.65; 0.79 | <0.001 | 92.9% |
| S.aureus | 26 | 682 | 0.74 | 0.70; 0.77 | 0.83 | 0.71; 0.82 | <0.001 | 90.4% |
| S. epidermidis | 20 | 956 | 0.69 | 0.66; 0.72 | 0.71 | 0.60; 0.80 | <0.001 | 88.9% |
| Other CNS | 26 | 906 | 0.72 | 0.68; 0.75 | 0.78 | 0.70; 0.86 | <0.001 | 77.1% |
| Total streptococci | 23 | 412 | 0.38 | 0.32; 0.43 | 0.50 | 0.32; 0.68 | <0.001 | 87.7% |
| S. pneumoniae | 19 | 212 | 0.31 | 0.23; 0.40 | 0.36 | 0.14; 0.60 | <0.001 | 79.3% |
| S. pyogenes | 15 | 52 | 0.76 | 0.58; 0.90 | 0.83 | 0.56; 1.00 | <0.001 | 61% |
| Other streptococci | 14 | 105 | 0.09 | 0.02; 0.18 | 0.19 | 0.03; 0.41 | <0.001 | 67.8% |
| S. agalactiae | 13 | 43 | 0.92 | 0.75; 1.00 | 0.90 | 0.71; 1.00 | 0.27 | 15.2% |
| Total enterococci | 24 | |||||||
| E. faecalis | 24 | 252 | 0.88 | 0.82; 0.93 | 0.86 | 0.74; 0.95 | <0.001 | 67.3% |
| E. faecium | 22 | 179 | 0.84 | 0.76; 0.91 | 0.83 | 0.69; 0.94 | <0.001 | 58.4% |
CNS, coagulase negative staphylococci.
A total of 27 studies, including 3904 samples, reported results for Gram-positive. The CIR in individual studies ranged from 0.17 to 0.98 at the species level. High heterogeneity was found in this meta-analysis (I2: 96.5%; p<0.001) (Fig. 3). The estimated pooled CIR was 0.72 (95% CI: 0.64–0.80) using the random-effects model. The funnel plot and Egger's test (p=0.06) showed that publication bias was not significant (Supplementary Fig. S2). A sensitivity analysis excluding the study 45 with a CIR of 0.17 (95% CI: 0.13–0.22) only reduced the heterogeneity to I2: 93.1 (p<0.001) and the pooled CIR estimated using a random effect model changed to 0.75 (95%CI:0.69–0.88). The rapid identification rates for different Gram-positive bacteria in blood cultures are recorded in Table 2. The rates of correct identification of Staphylococcus aureus, Staphylococcus epidermidis and other coagulase negative staphylococci were over 0.70. The results for streptococci were described in 23 studies and 412 reported samples. The CIRs of these pathogens, especially Streptococcus pneumoniae (0.36) and other alfa-hemolytic streptococci (0.19), were lower than those observed for staphylococci. The CIR of Streptococcus pyogenes was 0.83 and of Streptococcus agalactiae, 0.92. The rates of direct identification of Enterococcus faecalis and Enterococcus faecium were higher than 0.84 for both species. In these analyses, heterogeneity for each species of Gram-positive bacteria was significant (except for S. agalactiae), with I2 values above 60% (p<0.001), (Table 2).
The forest plot for the meta-analysis: identification of Enterobacteriaceae at the species level. CI, confidence interval; W, weight; fixed, fixed-effect model; random, random-effects model; events, number of correct identifications; total, total number of identifications. Gray squares represent the weight of individual studies with the fixed-effect model; horizontal lines through the squares represent 95% confidence intervals; gray diamonds represent the overall estimate and its confidence interval; dotted vertical lines represent the fixed-effect model; and dashed vertical lines represent the random-effects model.
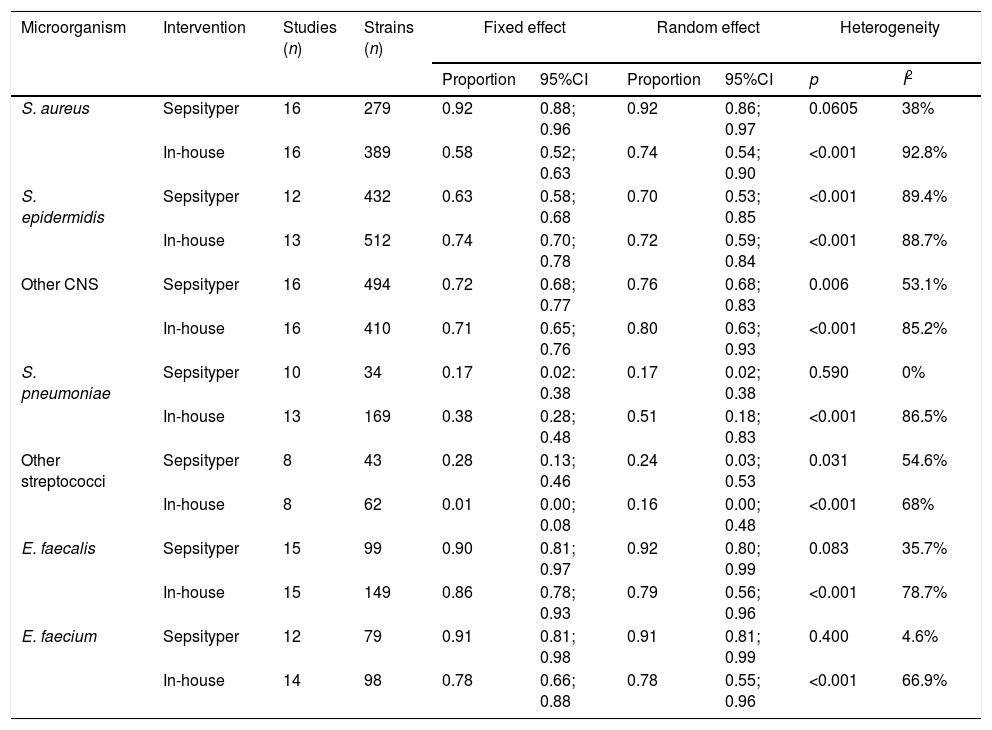
Sub-group analyses were performed to determine possible variables associated with the poorer rates of identification of Gram-positive bacteria (staphylococci, streptococci). For the sub-analyses, the studies were divided into those that used “in-house” methods for pretreatment of the sample and those that used the commercial Sepsityper method (Table 3). The commercial Sepsityper method gave higher percentages of correct identifications of S. aureus (0.92 vs. 0.58). S. pneumoniae, however, showed superior results with in-house methods, although the identification rate was low with both methods (0.51 vs. 0.17). The results for other staphylococci and streptococci were similar, with no differences between the two methods. In the case of enterococci, the results were slightly higher using the Sepsityper kit (0.92 vs. 0.78). There was lower heterogeneity in these sub-group analyses with the commercial Sepsityper method (I2 values ranged from 0 to 89.4%, but was only significant in one species, S. epidermidis) compared with in-house methods (I2 values ranged from 66.9% to 92.8% and significant in all species).
Differences between in-house methods and the Sepsityper method among Gram-positive bacteria.
| Microorganism | Intervention | Studies (n) | Strains (n) | Fixed effect | Random effect | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|---|
| Proportion | 95%CI | Proportion | 95%CI | p | I2 | ||||
| S. aureus | Sepsityper | 16 | 279 | 0.92 | 0.88; 0.96 | 0.92 | 0.86; 0.97 | 0.0605 | 38% |
| In-house | 16 | 389 | 0.58 | 0.52; 0.63 | 0.74 | 0.54; 0.90 | <0.001 | 92.8% | |
| S. epidermidis | Sepsityper | 12 | 432 | 0.63 | 0.58; 0.68 | 0.70 | 0.53; 0.85 | <0.001 | 89.4% |
| In-house | 13 | 512 | 0.74 | 0.70; 0.78 | 0.72 | 0.59; 0.84 | <0.001 | 88.7% | |
| Other CNS | Sepsityper | 16 | 494 | 0.72 | 0.68; 0.77 | 0.76 | 0.68; 0.83 | 0.006 | 53.1% |
| In-house | 16 | 410 | 0.71 | 0.65; 0.76 | 0.80 | 0.63; 0.93 | <0.001 | 85.2% | |
| S. pneumoniae | Sepsityper | 10 | 34 | 0.17 | 0.02: 0.38 | 0.17 | 0.02; 0.38 | 0.590 | 0% |
| In-house | 13 | 169 | 0.38 | 0.28; 0.48 | 0.51 | 0.18; 0.83 | <0.001 | 86.5% | |
| Other streptococci | Sepsityper | 8 | 43 | 0.28 | 0.13; 0.46 | 0.24 | 0.03; 0.53 | 0.031 | 54.6% |
| In-house | 8 | 62 | 0.01 | 0.00; 0.08 | 0.16 | 0.00; 0.48 | <0.001 | 68% | |
| E. faecalis | Sepsityper | 15 | 99 | 0.90 | 0.81; 0.97 | 0.92 | 0.80; 0.99 | 0.083 | 35.7% |
| In-house | 15 | 149 | 0.86 | 0.78; 0.93 | 0.79 | 0.56; 0.96 | <0.001 | 78.7% | |
| E. faecium | Sepsityper | 12 | 79 | 0.91 | 0.81; 0.98 | 0.91 | 0.81; 0.99 | 0.400 | 4.6% |
| In-house | 14 | 98 | 0.78 | 0.66; 0.88 | 0.78 | 0.55; 0.96 | <0.001 | 66.9% | |
This meta-analysis synthesizes the available evidence about the validity of the MALDI-TOF system for the accurate identification of bacteria directly from positive blood cultures. An assessment of the results suggests that MALDI-TOF provides highly accurate identification of Gram-negative bacteria at the species level directly from positive blood cultures. For Gram-positive bacteria, overall accuracy is moderate. Individual studies have previously evaluated the different percentages of correct identifications between Gram-negative and Gram-positive bacteria.12,41 This has several potential clinical implications. On the one hand, because the susceptibility patterns of some Gram-negative bacteria (e.g., P. aeruginosa, A. baumannii, some Enterobacteriaceae) are typically different, accurate identification is important and could therefore allow more rapid optimization of targeted therapy. On the other hand, the results found for Gram-positive are somewhat worrisome; the early management of BSI due to S. aureus and coagulase-negative staphylococci can be quite different, so that misidentification of these bacteria and other Gram-positive may have negative consequences for patient care.
It is not yet understood why it is not always possible to identify Gram-positive bacteria directly from positive blood cultures. It has been speculated that the more robust cell wall decreases protein extraction efficacy and that the slow growth of some species can lead to a very small pellet after the extraction. Several studies have shown that, in terms of efficacy (percentage of correct identifications compared to established reference methods), the Sepsityper performs best with Gram-negative bacteria. Nevertheless, even with Gram-positive bacteria, some researchers report successful identifications in around 75% or more of positive blood cultures.48
Various studies have been published comparing the results obtained with the Sepsityper system and different in-house methods before carrying out the MALDI-TOF analysis. However, while some studies have demonstrated that alternative extraction methods may yield results identical to or better than the Sepsityper kit,6,10,41 others have demonstrated the superiority of the Sepsityper extraction method over in-house methods.16,46 To determine the possible advantage of the Sepsityper kit for the correct identification of Gram-positive bacteria, we analyzed separately those studies that had used in-house methods for pretreatment of the sample and those that had used the commercial Sepsityper method. Although the Sepsityper method showed higher identification rates for some species, the in-house methods generally generated similar results to those obtained with Sepsityper.
Overall, our results showed no major differences in the identification of Gram-positive bacteria when the Sepsityper method or in-house methods were used, although the heterogeneity of results arising from studies that used the Sepsityper method was lower than when in-house methods were used. Nevertheless, the rate of correct identifications of S. aureus was clearly higher with the Sepsityper method, and S. pneumoniae showed superior results with the in-house, as against the Sepsityper method. The results for other Staphylococcus spp. and Streptococcus spp. were similar, with no differences between the two methods. For the identification of Enterococcus spp., the results recorded were slightly higher using the Sepsityper, compared to in-house methods. As with previous reports, our results suggest that the decision to implement one or the other depends on the individual laboratory.
In most cases, the methodological quality of the studies included in this review is moderate. The studies have various limitations as well as methodological issues, with problems of both internal and external validity. Some of these limitations were: the selection of samples included in the different studies; lack of blinding; both tests in the included studies were not realized simultaneously; the intervention differed depending on whether the method used was in-house or commercial; likewise, the subsequent interpretation of the data varied as a result of the different cut-offs used; finally the microbiological reference method in the included studies was performed differently, using different techniques and procedures.
To the best of our knowledge, this is the first meta-analysis of studies of diagnostic tests to assess the accuracy of MALDI-TOF for the rapid, direct identification of bacteria from positive blood cultures. Earlier studies have evaluated the MALDI-TOF system for the identification of clinical bacteria; Drancourt49 published a review of MALDI-TOF for the detection of microorganisms in the blood; Morgenthaler et al.48 reviewed only the standard Sepsityper method for rapid identification of microorganisms, including in-house methods, although the review was not systematic. In 2015, Dixon et al.50 carried out a systematic review comparing MALDI-TOF and conventional identification methods for the rapid identification of pathogens in patients with suspected or known BSI, and also collected information on reduced hospital costs, length of stay, and ‘time to appropriate antimicrobial treatment’. Ling et al.,51 in 2014, published a meta-analysis of the accuracy of MALDI-TOF for the identification of clinical pathogenic fungi, but not bacteria.
This meta-analysis has a number of limitations. First, the number and heterogeneity of the studies posed a variety of research questions. A second limitation was potential publication bias; we excluded unpublished papers, gray literature and industry reports. We tried to avoid this bias as much as possible by using several databases and performing searches without language restrictions. In order to avoid possible bias in the application of selection criteria, these were pre-specified a priori. A third limitation is that some studies conducted the test with two or three different interventions (e.g.: Sepsityper and one or two in-house methods). We assumed that each intervention was a new independent study, so that the total number of evaluated strains was higher. A fourth limitation is that some studies presented score cut-off values higher than 2.0 for the MALDI-TOF detection system. We assumed that all results had a cutoff point of ≥2.0 to try and ensure standardization of the results. Another possible limitation was that, although all studies used an adequate gold standard as comparator, these varied across the studies, so that the results would have varied depending on the quality of the gold standard. The results of this study therefore should be interpreted with caution due to these potential limitations. With respect to the high heterogeneity obtained for overall Gram-negative bacteria in the meta-analysis, this may be related to the inclusion of different species where the identification accuracy of the MALDI-TOF system was heterogeneous, since heterogeneity decreased substantially when the accuracy of each specific species of Gram-negative bacteria was analyzed separately, as previously stated. On the other hand, the main reason for high heterogeneity among Gram-positive bacteria could be the inclusion of the two methods (both in-house or commercial) in the same overall meta-analysis, since it was previously confirmed in our separate analyses that heterogeneity for each Gram-positive species was higher with in-house methods. Another possible limitation of our work has been that from the end of it until its possible publication, new articles of interest have been published in the scientific literature, which could partially modify the results of our systematic review.
Finally, this meta-analysis highlights the need to perform new diagnostic studies to assess the accuracy of MALDI-TOF for Gram-positive bacteria with standardized methods, using the same comparator to improve the methodology. A faster time-to-result is an advantage for the patient, since, in combination with antibiotic stewardship programs, optimized antibiotic therapy can very often be administered on the basis of a species/genus identification of the underlying microorganisms. Looking to the future, we may suppose that this technology can be used in the detection of bacterial resistance determinants and virulence factors.
In summary, MALDI-TOF MS demonstrated high accuracy for the direct identification of Gram-negative bacteria from blood culture and moderate accuracy for Gram-positive bacteria (low in some species). There were no major differences with respect to the identification of Gram-positive bacteria when either in-house methods or the standard Sepsityper kit were used.
Conflict of interestsAll authors confirm that there is no conflict of interest for the publication of this work.