We have developed a MALDI-TOF-mediated phenotypic method, which determines antibiotic susceptibility (AS) from positive blood cultures (BCs) in 2h. We developed a software for process automation. We report results on Escherichia coli-positive BCs with cefotaxime (CTX) and ciprofloxacin (CIP).
MethodsWe studied CIP and CTX activity in 18 and 17 real E. coli-positive BCs, and in 56 and 45 spiked BCs, respectively. Positive BCs were incubated for 2h without any antibiotics, and with 2mg/l and 4mg/l of CIP and CTX. The extraction was performed using ethanol/formic acid. Spectra were processed with specifically developed software which compares the peaks’ intensity and the size of specific peaks.
ResultsThe set cut-off point was a 3-fold decrease in the summation of all peaks and/or the 5382m/z peak value (ribosomal protein L34). In simulated BCs, the correlation of CIP 2mg/l and 4mg/l with Etest® was 94.6% and 98.2%, respectively; for CTX 2mg/l and 4mg/l, this correlation was 95.6%. In real BCs, the correlations were 100% for CIP (2mg/l and 4mg/l) and 88.2% and 94.1% for CTX 2mg/l and 4mg/l, respectively. Resistant isolates were always correctly classified.
ConclusionThis method provides accurate, fast and inexpensive AS information. The method can be automated, making it easier to implement in a microbiology laboratory routine.
Se ha desarrollado un método fenotípico basado en MALDI-TOF, que determina la sensibilidad a antibióticos en hemocultivos (HC) positivos en 2h. Se ha desarrollado un software que automatiza el proceso. Se presentan los resultados en HC positivos para Escherichia coli, con cefotaxima (CTX) y ciprofloxacino (CIP).
MétodosSe estudió la actividad de CIP y CTX en 18 y 17HC positivos reales con E. coli, y en 56 y 45 HC simulados. Los HC positivos se incubaron durante 2h sin antibiótico, y con 2 y 4 mg/l de CIP y de CTX. La extracción se realizó con etanol/ácido fórmico. Los espectros se procesaron con un software específico, que compara la intensidad de los picos y el tamaño de los picos específicos.
ResultadosEl punto de corte establecido fue una disminución de 3 veces en la suma de picos, y/o en el valor del pico de 5.382 m/z (proteína ribosómica L34). En hemocultivos simulados la correlación con Etest® para las concentraciones de CIP de 2 y 4mg/l fueron 94,6 y 98,2%, respectivamente, y 95,6% para CTX (2 y 4mg/l). En HC reales, la correlación con Etest® fue del 100% para CIP (2 y 4mg/l), y del 88,2 y 94,1% para CTX 2 y 4mg/l, respectivamente. Los aislados resistentes siempre se clasificaron correctamente.
ConclusiónEste método proporciona información sobre sensibilidad a antimicrobianos de manera precisa, rápida y barata. El método se puede automatizar e incluir en la rutina del laboratorio de microbiología.
MALDI-TOF MS is a fast and reliable method, which allows microorganism identification hours, or even days sooner than conventional methodology.1–3 MALDI-TOF MS has been shown a very accurate method when working from colonies,4 but also from blood cultures (BC)5,6 and even directly from some samples.7 Nevertheless, the time saved in identification frequently cannot be fully translated into clinical efficiency, because antimicrobial susceptibility methods cannot offer a similar agility. Getting, through MALDI-TOF MS, an antibiogram offering the same information as conventional antibiograms, and the speed and reliability of MALDI-TOF MS identifications, is a major challenge. Most antibiotics can become inactive through a number of resistance mechanisms, and a great diversity of proteins, in terms of size, production level, cellular location, etc., can be involved. Methods allowing to detect the presence of enzymes hydrolyzing certain antibiotics, or the presence of certain resistance mechanisms, have been described.8–11 These strategies can eventually provide useful and rapid information concerning a particular group of antibiotics, but we cannot expect to obtain global susceptibility profiles through these methods. New strategies, such as the incorporation into the culture medium of labeled aminoacids,12 or quantitative MALDI-TOF MS have been developed.13–15
We have developed a MALDI-TOF MS-based susceptibility method, based on the behavior of specific MALDI-TOF MS peaks in the presence and absence of antibiotics. Though the method can be applied manually to the information reported by the FlexAnalysisR software (Bruker Daltonics GmbH, Germany), we have also developed a specific software which allows process automation. Our method offers phenotypic information on antibiotic susceptibility, regardless of the resistance mechanisms involved, in 2h, and can be directly applied to positive BC.
The aim of this study was to test the sensitivity and specificity of this method with real and spiked BC positive for Escherichia coli for ciprofloxacin (CIP) and cefotaxime (CTX).
Material and methodsWe studied 35 real E. coli-positive BCs and 101 spiked BCs inoculated with E. coli ATCC 25922 (2 BCs) and with 99 recent clinical isolates of E. coli, as previously described.6 Shortly, three to four 24h old E. coli colonies grown on Mueller Hinton agar were suspended into sterile water to reach a turbidity of 0.5 in the McFarland scale. The E. coli suspension was diluted 1/10 in sterile water, and 1mL of this suspension was inoculated into BACTEC Plus+Aerobic/F bottles (Becton Dickinson, NJ, USA), and incubated at 37°C in a BACTEC 9240 device (Becton Dickinson, NJ, USA), until they were reported as positive.
Real BCs were processed according to manufacturer's instructions and incubated at 37°C in a BACTEC 9240 device (Becton Dickinson, NJ, USA).
Both the isolates inoculated and the isolates obtained from real BCs were studied for antibiotic susceptibility by microdilution (Vitek 2, bioMérieux, France). MICs were confirmed by Etest® (bioMérieux, France), following CLSI guidelines.16
Once reported as positive, both simulated and real BCs were randomly divided into two sets: the first set (18 real BCs and 56 spiked BCs, including one inoculated with E. coli ATCC 25922) were studied for MALDI-TOF MS-mediated CIP susceptibility.
The second set (17 real BCs and 45 spiked BCs, including one inoculated with E. coli ATCC 25922) were studied for MALDI-TOF MS-mediated CTX susceptibility.
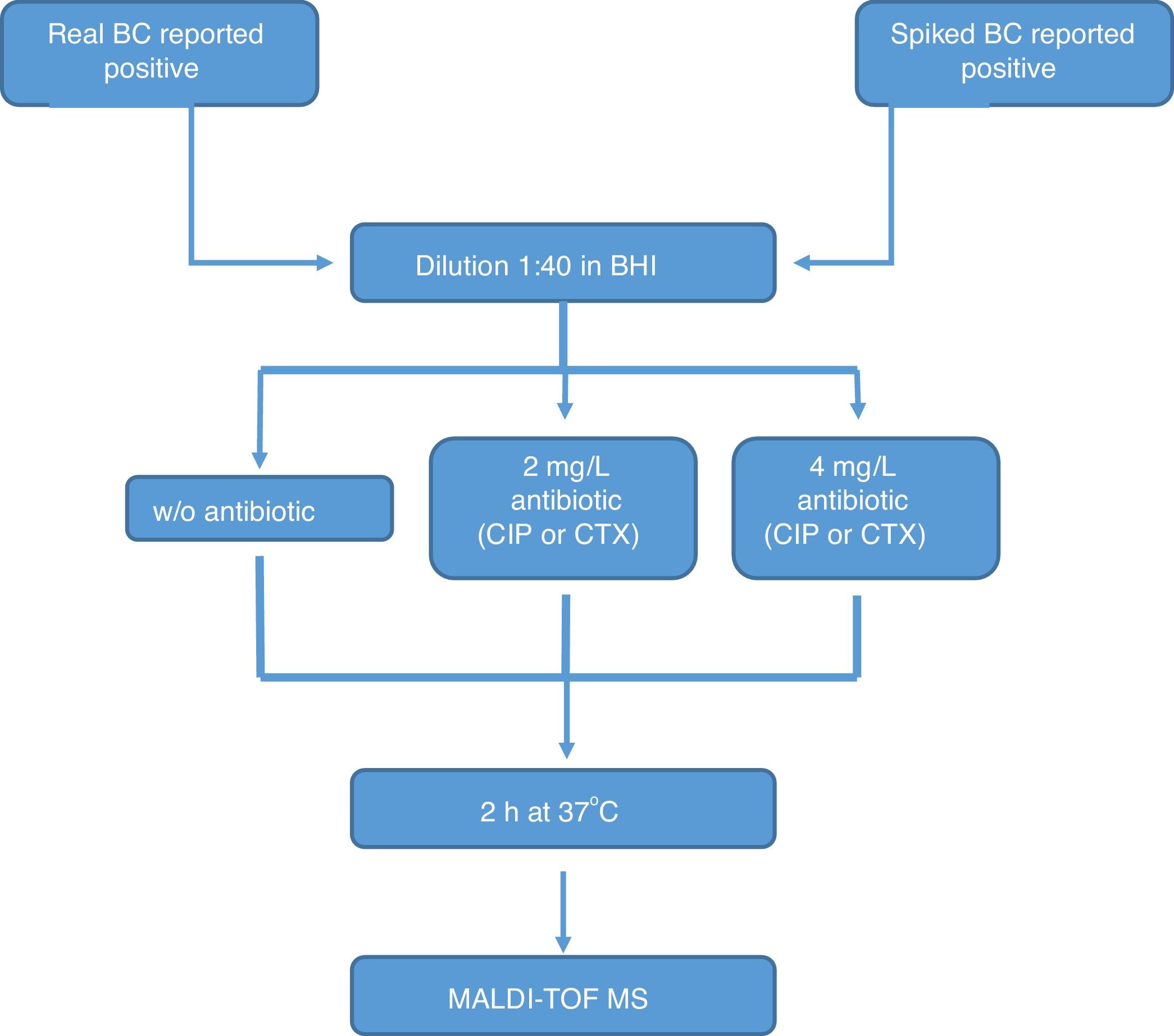
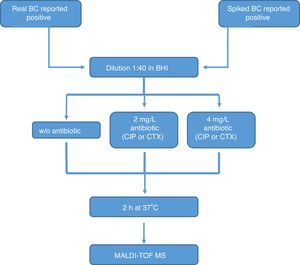
All the bottles (both sets) were processed as follows (Fig. 1): (1) tube A: 10μl of BC were diluted in 390μl of BHI broth and incubated at 37°C for 2h; (2) tube B: 10μl of BC were diluted in 390μl of BHI broth with CIP (1st set) or CTX (2nd set) at 2mg/l and incubated at 37°C for 2h; (3) tube C: 10μl of BC were diluted in 390μl of BHI broth with CIP (1st set) or CTX (2nd set) at 4mg/l and incubated at 37°C for 2h. After incubation, we processed the BCs as described by Lange et al.,13 with some modifications. Briefly, cells were centrifuged and washed once with 150μl pure water and once with 200μl 70% ethanol. Then, bacteria were lysed according to the MALDI BiotyperR workflow using an ethanol/formic acid extraction. One μl of lysate was directly spotted in a ground steel MALDI target plate (Bruker Daltonics GmbH, Germany). Each lysate was spotted twice.
MALDI-TOF mass spectrometry was performed as previously described,6 on an Autoflex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics GmbH, Germany) equipped with a 200-Hz Smartbeam laser.
Data analysis. For automated data analysis, raw spectra were processed using the MALDI BiotyperR 3.0 software (Bruker Daltonics GmbH, Germany) at default settings. The software performs normalization, smoothing, baseline subtraction, and peak picking, creating a list of the most significant peaks of the spectrum (m/z values with a given intensity, with the threshold set to a minimum of 1% of the highest peak and a maximum of 100 peaks). MALDI-TOF MS identifications were classified as follows: a score ≥2 indicates species identification; a score between 1.7 and 1.9 indicates genus identification, and a score <1.7 indicates no identification.
Antibiotic resistance. Spectra generated by MALDI BiotyperR were processed in FlexAnalysisR software (Bruker Daltonics GmbH, Germany). A specific processing method was used in order to detect as many peaks as possible, even though they were not relevant for the identification process. The spectra were first baseline substrated (TopHat algorithm), normalized and smoothed (Savitzkty Golay algorithm). The signal to noise threshold was 0.001 and the peak detection algorithm chosen was centroid with a maximum of 150 peaks.
Once the full protein profiles of each sample were obtained from FlexAnalysisR, the files containing all this information were processed with a software specifically developed. The system has been developed with the NET framework using the C# language. It consists of a tool that facilitates the integrated management and visualization of all the data on the intensities of the m/z peaks collected for each strain. This software only had in account m/z peaks with a value ≥2500. In its first version, the system consists of four main modules: (1) module for peaks data import and integration which, based on regular expressions, identifies the XML files that contain the data obtained for a particular isolate, and incorporates them in an integrated way in a new structure, which can be stored in a new XML file; (2) module for data homogenization: the values obtained for each m/z peak in the different profiles of the same isolate are analyzed, establishing the clusters that will be considered representative of each protein. This strategy allows further comparisons and visual alignments between different profiles. The clustering strategy was agglomerative, based on Euclidean distance. (c) visualization: this module allows a graphic representation of data, making easier their visual analysis. The software includes some tools allowing to configure the elements that are being included in the graph: (d) extraction of relevant data: from the data of a set of strains new data structures are generated with the information that has been considered of interest for the study.
This software obtains the whole peaks profile from FlexAnalysisR processed spectra, and compares the summation of the intensities of the full group of peaks of each profile, and the size of specific peaks in each profile, having mainly in account the proportion between the summation of all the peaks in the tubes with antibiotics and the tube without antibiotic after a 2-h incubation, and the proportion between the 5382m/z peak, which represents the ribosomal protein L34, and is the most deeply and rapidly affected by the action of the antibiotic, in the tubes with antibiotics and the tube without antibiotic after a 2-h incubation.
The main criteria used were a 3-fold decrease in the summation of the peaks obtained, and/or a 3-fold decrease in the 5382m/z peak value in the tubes with antibiotic, with respect to the tube grown for 2h without antibiotic. This comparison was also made manually. When no growth was found in any tube, including the control grown without antibiotic, the test was considered invalid. The whole algorithm for our method is shown in Fig. 1.
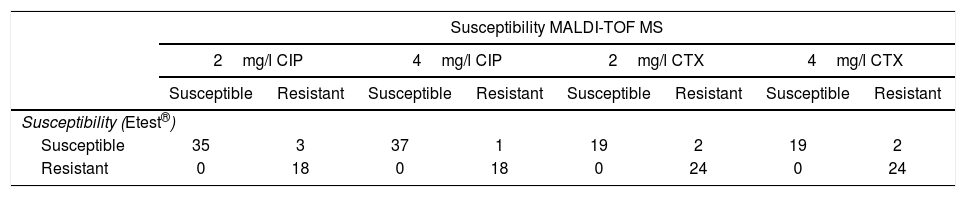
ResultsMICs of CIP in CIP-susceptible and CIP-resistant isolates used for spiked BC ranged between ≤0.008 and 1mg/l, and 2 and >32mg/l respectively. MICs of CIP of E. coli isolates obtained from real BC ranged between 0.003mg/l and >32mg/l. Results for spiked BC appear in Table 1. 2mg/l CIP tubes correlated with Etest® in 94.6% of cases (53/56). All the resistant isolates and 92.1% of susceptible isolates were classified correctly by our method, while 3 susceptible isolates (MICs of CIP by Etest®: 0.25, 0.25 and 1mg/l) were classified as resistant. The 18 isolates resistant by Etest® harbored gyrA or gyrA+parC mutations.
Results obtained with the method proposed, in comparison with Etest®, in spiked blood cultures with E. coli, for ciprofloxacin (CIP) and cefotaxime (CTX).
| Susceptibility MALDI-TOF MS | ||||||||
|---|---|---|---|---|---|---|---|---|
| 2mg/l CIP | 4mg/l CIP | 2mg/l CTX | 4mg/l CTX | |||||
| Susceptible | Resistant | Susceptible | Resistant | Susceptible | Resistant | Susceptible | Resistant | |
| Susceptibility (Etest®) | ||||||||
| Susceptible | 35 | 3 | 37 | 1 | 19 | 2 | 19 | 2 |
| Resistant | 0 | 18 | 0 | 18 | 0 | 24 | 0 | 24 |
Using the same parameters, 4mg/l CIP tubes correlated with Etest® results in 98.2% of cases (55/56). All the resistant isolates and 97.4% of susceptible isolates were classified correctly by our method. One susceptible isolate (MIC of CIP by Etest®: 1mg/l) was classified as resistant.
The same parameters as in spiked BC were used for real BC. Results both with CIP concentrations of 2mg/l and 4mg/l correlated with Etest® results in 100% of cases (18/18) (Table 2). As in spiked blood cultures, the 8 CIP-resistant isolates by Etest® harbored gyrA or gyrA+parC mutations.
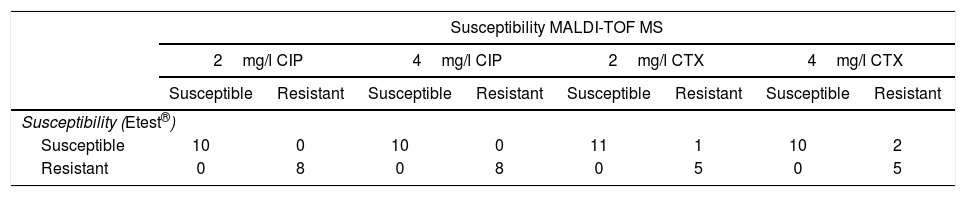
Results obtained with the method proposed, in comparison with Etest®, in real blood cultures with E. coli, for ciprofloxacin (CIP) and cefotaxime (CTX).
| Susceptibility MALDI-TOF MS | ||||||||
|---|---|---|---|---|---|---|---|---|
| 2mg/l CIP | 4mg/l CIP | 2mg/l CTX | 4mg/l CTX | |||||
| Susceptible | Resistant | Susceptible | Resistant | Susceptible | Resistant | Susceptible | Resistant | |
| Susceptibility (Etest®) | ||||||||
| Susceptible | 10 | 0 | 10 | 0 | 11 | 1 | 10 | 2 |
| Resistant | 0 | 8 | 0 | 8 | 0 | 5 | 0 | 5 |
Concerning isolates used for comparing CTX susceptibility, 21 CTX-susceptible and 24 CTX-resistant E. coli isolates were used for spiked BC. All CTX-resistant isolates were ESBL producers. No carbapenemase-producing isolates were tested. 12 E. coli isolates obtained from real BC were CTX-susceptible and 5 isolates were CTX-resistant. These 5 CTX-resistant isolates were also ESBL-producers.
Results concerning spiked BC are shown in Table 1. Using the same criteria enounced for CIP, the results obtained both with 2 and 4mg/l CTX concentrations correlated with Etest® in 95.6% of cases (43/45). All the resistant isolates and 90.5% of susceptible isolates were classified correctly by our method, while 2 susceptible isolates (MICs of CTX by Etest®: 1 and 0.06mg/l) were classified as resistant.
Results for real BC with CTX concentrations of 4mg/l correlated with Etest® in 94.1% of cases (16/17) (Table 2). All the resistant isolates were classified correctly and 8.3% of susceptible isolates (1/12) (MIC of CTX by Etest®: 1mg/l) were classified as resistant by our method. When CTX concentrations of 2mg/l were used, correlation with Etest® was 88.2% (15/17) (Table 2). All the resistant isolates were classified correctly. 16.7% of susceptible isolates (2/12) (MIC of CTX by Etest®: 0.06 and 1mg/l) were erroneously classified as CTX-resistant.
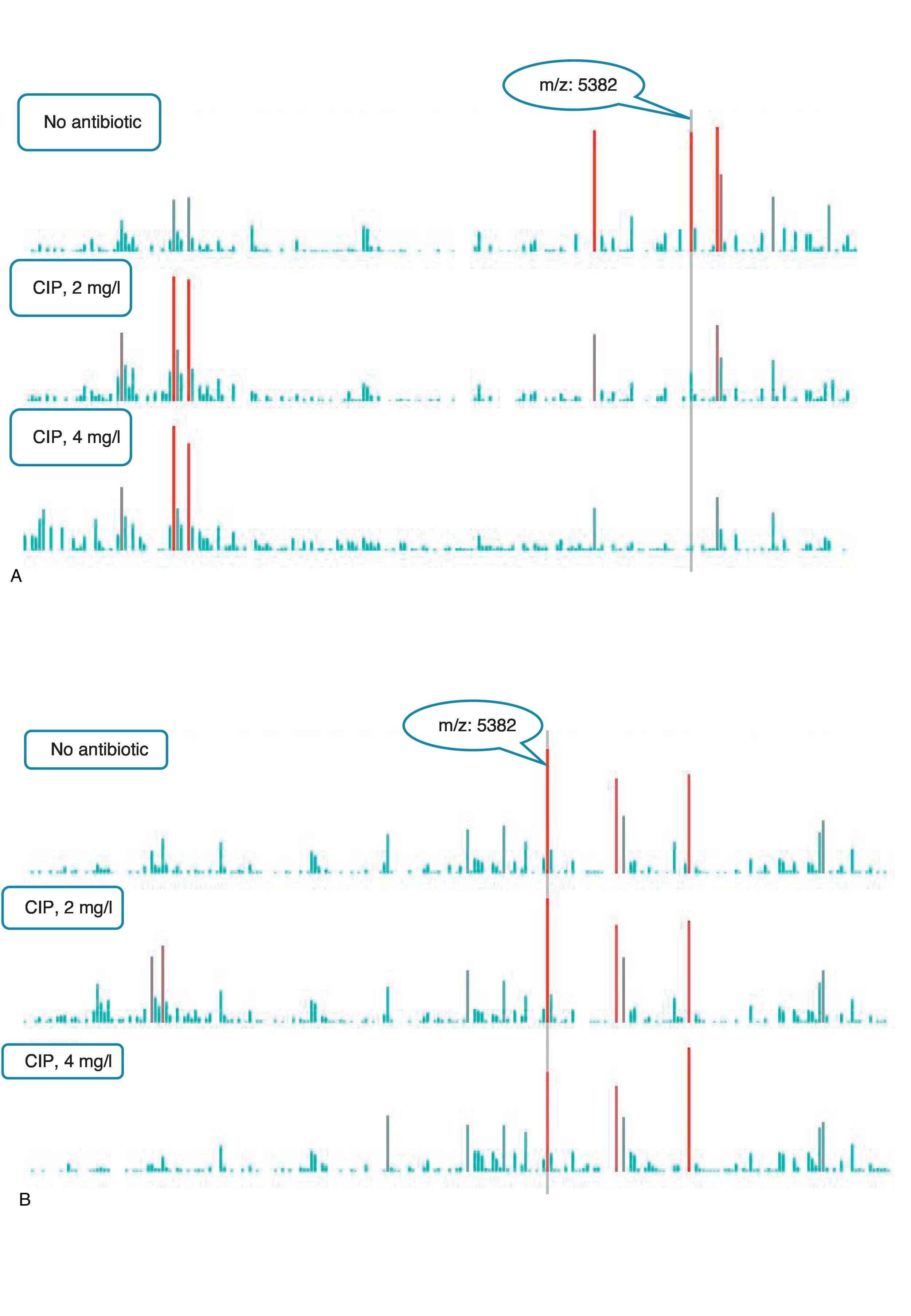
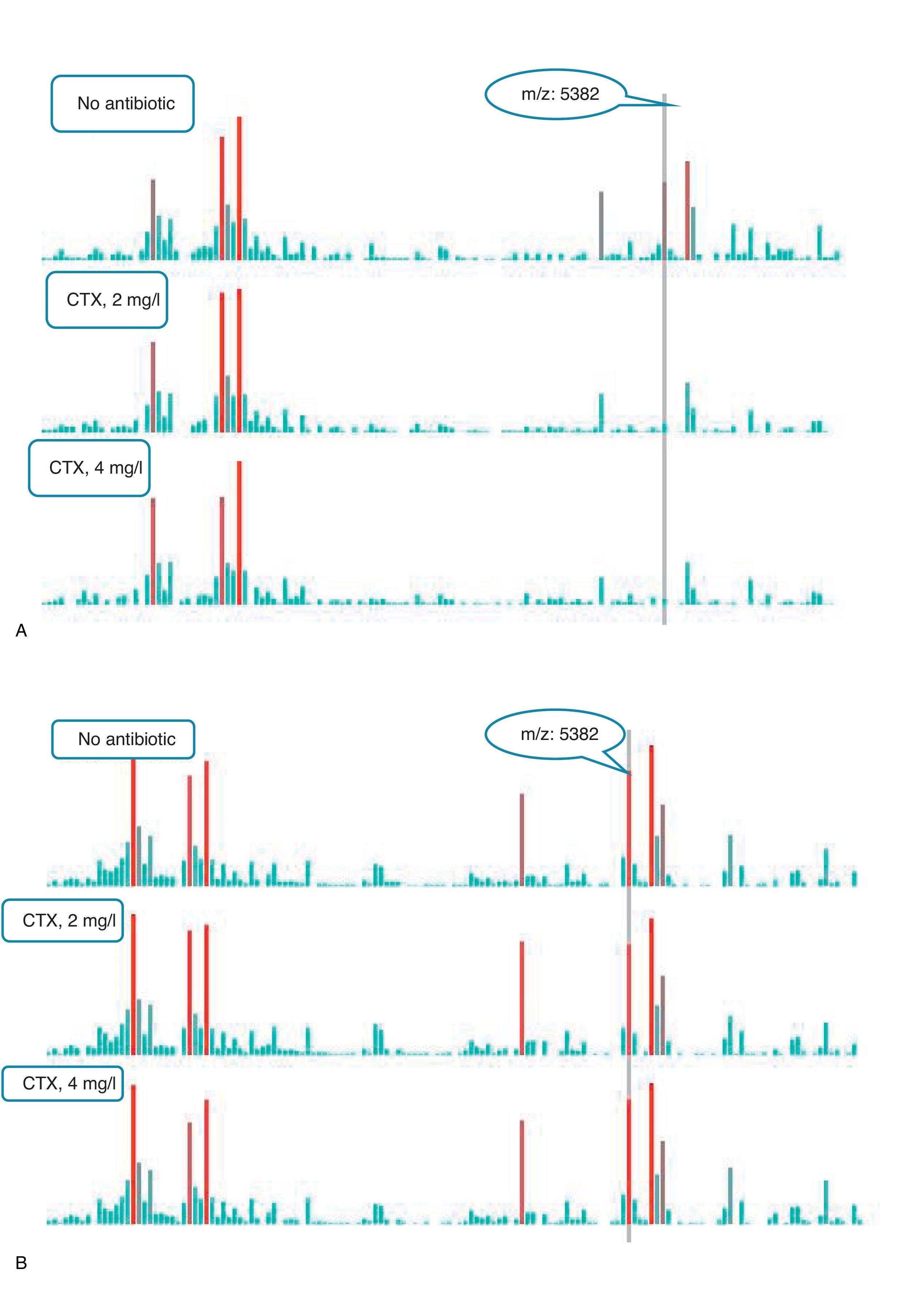
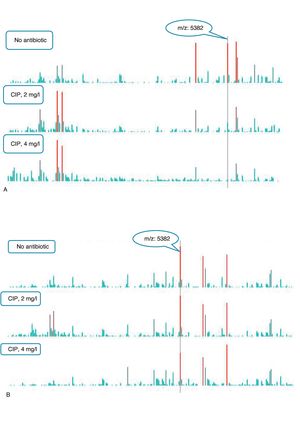
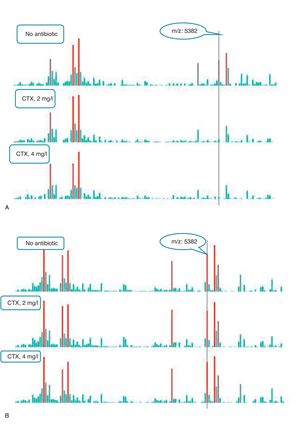
An example of graphics generated by the software from CIP-susceptible and CIP-resistant, and CTX-susceptible and CTX-resistant E. coli-positive blood cultures and the main m/z peaks used for susceptibility/resistance evaluation in shown in Figs. 2 and 3.
m/z peak's profiles obtained from blood cultures positive for ciprofloxacin-resistant and ciprofloxacin-susceptible E. coli, and main peaks used for categorization. (a) Blood culture positive for a ciprofloxacin- resistant E. coli. (b) Blood culture positive for a ciprofloxacin-susceptible E. coli.
m/z peak's profiles obtained from blood cultures positive for cefotaxime-resistant and cefotaxime-susceptible E. coli, and main peaks used for categorization. (a) Blood culture positive for a cefotaxime-resistant E. coli. (b) Blood culture positive for a cefotaxime- susceptible E. coli.
A pending subject for MALDI-TOF MS in clinical microbiology has been its application to the determination of antimicrobial susceptibility. Obtaining, through proteomic techniques, susceptibility profiles similar to those provided by conventional bacteriological techniques, with a speed and reliability close to those achieved in identification, is a major challenge, because of the great variety of resistance mechanisms, and the great diversity of proteins associated to them.
Methods have been developed allowing to detect some antibiotic-hydrolyzing enzymes (ESBLs, carbapenemases, etc.) based on the different m/z peaks generated by the intact antimicrobial molecule and the hydrolyzed molecule.8,9 Other resistance mechanisms, such as vanB, can be also deduced from the presence of specific peaks.10 A more recent paper has been published in which authors use a similar strategy for detecting fluoroquinolone resistance mediated by a specific mechanism.11 In this paper, the authors detected the presence of a fluoroquinolone-inactivating enzyme on the basis of the different m/z rate of the antibiotic and the antibiotic acetylated by the enzyme AAC-(6′)-Ib-cr.
These strategies can eventually provide a useful and rapid information regarding a particular mechanism of resistance or a particular group of antimicrobial agents, but we cannot expect to obtain, through these strategies, global susceptibility profiles comparable to those obtained with conventional antibiograms. Antimicrobial resistance is frequently associated, in whole or in part, to non-hydrolytic mechanisms, so that this methodology can allow to predict resistance, but in no case susceptibility. Moreover, in the case of some resistance mechanisms such as ESBLs, the current tendency is not to consider resistance if specific MIC values are not reached, even if the enzyme is present, so that the mere detection of hydrolysis would not be enough to issue a resistance report.
These limitations have stimulated the development of new strategies. A new methodology12 suggests the incorporation of isotopically labeled amino acids into the culture medium, along with the antibiotic to be tested. Susceptible microorganisms replication will be slower, and therefore will hardly incorporate labeled amino acids, while resistant microorganisms will multiply in a much more active way, and will incorporate larger amounts of labeled amino acids. These labeled aminoacids have specific sizes, and then will generate specific m/z profiles in resistant microorganisms. This method has been shown useful for the identification of MRSA, and in the detection of resistance to beta-lactams, aminoglycosides and fluoroquinolones. Another recently proposed alternative is semiquantitative MALDI-TOF mass spectrometry.13–15 This methodology is also based on the incubation of the microorganism in the presence and absence of the antimicrobials to be tested, and the quantification of bacterial growth in both circumstances. This method was initially tested with carbapenemase-producing Klebsiella spp.13, and has recently been tested with other microorganisms against several antibiotics,14 and even for mycobacteria,17 with good results. In this method,14 bacterial cells are extracted from the blood culture bottles, and used to prepare a bacterial suspension equivalent to around 5×106CFU/mL. We avoided this step because we had observed previously that, in enterobacteria, bacterial concentration at BC positivization is around 106CFU/mL. Our method does not loose specificity, especially in detecting resistance, and enhances simplicity and speed.
Our method is also based on the peak profile modifications conditioned by the growth of the microorganism in contact with specific antibiotic concentrations. The summation of peak values that we use is conceptually similar to the AUC used by these authors,14 and the relative growth (RG) defined by them is also conceptually similar to the rates between peaks values summations used by us. Moreover, we have found that the peak corresponding to the L34 ribosomal protein (5382m/z for E. coli) specifically drops in a fast and significant way in most antibiotic-susceptible microorganisms, and can be a helpful variable for predicting resistance. Moreover, the m/z value of this protein is well-known for most microorganisms, thus the search for this protein once the bacteria identification is allowed can be easily automated. This possible role of L34 protein as an early marker of slow growth has not been described previously, and seems to be associated to no specific mechanisms of action, since its behavior is similar for antibiotics with different mechanisms of action, such as fluoroquinolones and cephalosporins.
In whole, this method allows to predict the antibiotic susceptibility or resistance of the microorganisms, directly from the blood culture bottle, after 2h of incubation. Obviously the methods has, at this moment, different limitations. Previous publications18 show technical and biological variability when MALDI-TOF has been used for typing microorganisms. This aspect should not affect in the same extent to this method, since here we always compare profiles with and without antibiotics, obtained simultaneously and in the same conditions, but the reproductibility of the method shall be checked. Otherwise, we include in this paper only blood cultures positive for E. coli. The results with other genera and species shall be tested, and preliminar results with other microorganisms suggest a good behavior, at least with other Gram-negative microorganisms (unpublished data). The method provides susceptibility information in an accurate, fast and cheap way. Since broth inoculation and incubation with and without antibiotic are, in general terms, methodologically similar to broth microdilution susceptibility methods, this part might be susceptible of automation, making easier its implementation in the microbiology laboratory routine.
FundingThis study has been financed by the grant GRS 1213/A/15 (Gerencia Regional de Salud, Consejería de Sanidad, Junta de Castilla y León, Spain).
Conflicts of interestThe authors have no conflicts of interest to declare.