The discovery, commercialization and administration of antibiotics revolutionized the world of medicine in the middle of the last century, generating a significant change in the therapeutic paradigm of the infectious diseases. Nevertheless, this great breakthrough was soon threatened due to the enormous adaptive ability that bacteria have, through which they are able to develop or acquire different mechanisms that allow them to survive the exposure to antibiotics. We are faced with a complex, multifactorial and inevitable but potentially manageable threat. To fight against it, a global and multidisciplinary approach is necessary, based on the support, guidance and training of the next generation of professionals. Nevertheless, the information published regarding the resistance mechanisms to antibiotics are abundant, varied and, unfortunately, not always well structured. The objective of this review is to structure the, in our opinion, most relevant and novel information regarding the mechanisms of resistance to antibiotics that has been published from January 2014 to September 2019, analysing their possible clinical and epidemiological impact.
El descubrimiento, la comercialización y la administración de antibióticos revolucionó la medicina a mediados del siglo pasado, generando un cambio significativo en el paradigma terapéutico de las enfermedades infecciosas. Sin embargo, este avance no tardó en verse amenazado debido a la enorme capacidad que tienen las bacterias para desarrollar o adquirir distintos mecanismos que les permiten sobrevivir a los antibióticos. Nos encontramos frente a una amenaza compleja, multifactorial e inevitable, pero potencialmente manejable. Para luchar contra ella es necesario un abordaje multidisciplinar basado en el apoyo, la orientación y el entrenamiento de la próxima generación de profesionales. No obstante, la información publicada referente a nuevos mecanismos de resistencia a antibióticos es abundante, variada y, desgraciadamente, no siempre bien estructurada. El objetivo de esta revisión es ordenar la información, a nuestro juicio más relevante y novedosa, que se ha publicado en referencia los nuevos mecanismos de resistencia a los antibióticos desde enero de 2014 hasta septiembre de 2019, analizando su posible impacto clínico y epidemiológico.
The discovery, commercialization and administration of antibiotics revolutionized the world of medicine in the middle of the last century, generating a significant change in the therapeutic paradigm of the infectious diseases and giving an essential prophylactic coverage rate for the development of new surgical techniques, transplants and oncological treatments.
Nevertheless, this great breakthrough was soon threatened due to the enormous adaptive ability that bacteria have, through which they are able to develop or acquire different mechanisms that allow them to survive the exposure to antibiotics. Currently, the fight against antibiotic resistance is considered a health priority by the main national institutions (Ministry of Health, Autonomous Communities in Spain) and international [Centers for Disease Control and Prevention (CDC), European Center for Disease Prevention and Control (ECDC), World Health Organization (WHO)]. WHO considers antibiotic resistance one of the three main health threats of the 21st century worldwide.
The immense dynamism of bacterial biology, due to its genetic plasticity and high reproduction rate, encourages the continued emergence and selection of new mechanisms of antibiotic resistance as a response to the selective pressure implied in the use and abuse of these drugs, and therefore the growing emergence of bacteria with multiple resistance (MDR), and even extensive resistance (XDR) and pan-resistance (PDR) to antibiotics.1
Infections caused by resistant bacteria have a significant impact on the morbidity and mortality of patients, and generate a significant economic cost over those caused by susceptible bacteria.2 The dissemination of MDR and XDR bacteria, together with the slow pace of commercialization of new antibiotics, is generating a significant limitation in therapeutic options against the infections they produce.
We are faced with a complex, multifactorial and inevitable but potentially manageable threat.3 To fight against it, a global and multidisciplinary approach is necessary, which is currently being promoted by the different international health agencies and national governments through the creation of action plans to combat antimicrobial resistance. These initiatives must be based, among other aspects, on the support, guidance and training of the next generation of professionals focused on a rapid and accurate detection of resistant bacteria, of the mechanisms by which they are able to resist, and by which they spread.3 Undeniably, such knowledge and early detection should be reflected in the formulation of prevention and control strategies for the spread of these pathogens.
An increasing number of studies are aimed at detecting new mechanisms of resistance; the different lines of research look for variants of those already known, associations of more than one mechanism, mechanisms already described in other bacterial species and the emergence of new ones. Therefore, the information published in recent years regarding these issues is abundant, varied and, unfortunately, not always well structured.
The objective of this review is to structure the novel and relevant information that has been published in recent years on antibiotic resistance mechanisms, analyzing their possible clinical and epidemiological impact. To this end, the publications available in PubMed from January 2014 to September 2019 have been reviewed. The most relevant innovations in our opinion are summarized below and in Tables 1 and 2, sorted by antibiotics.
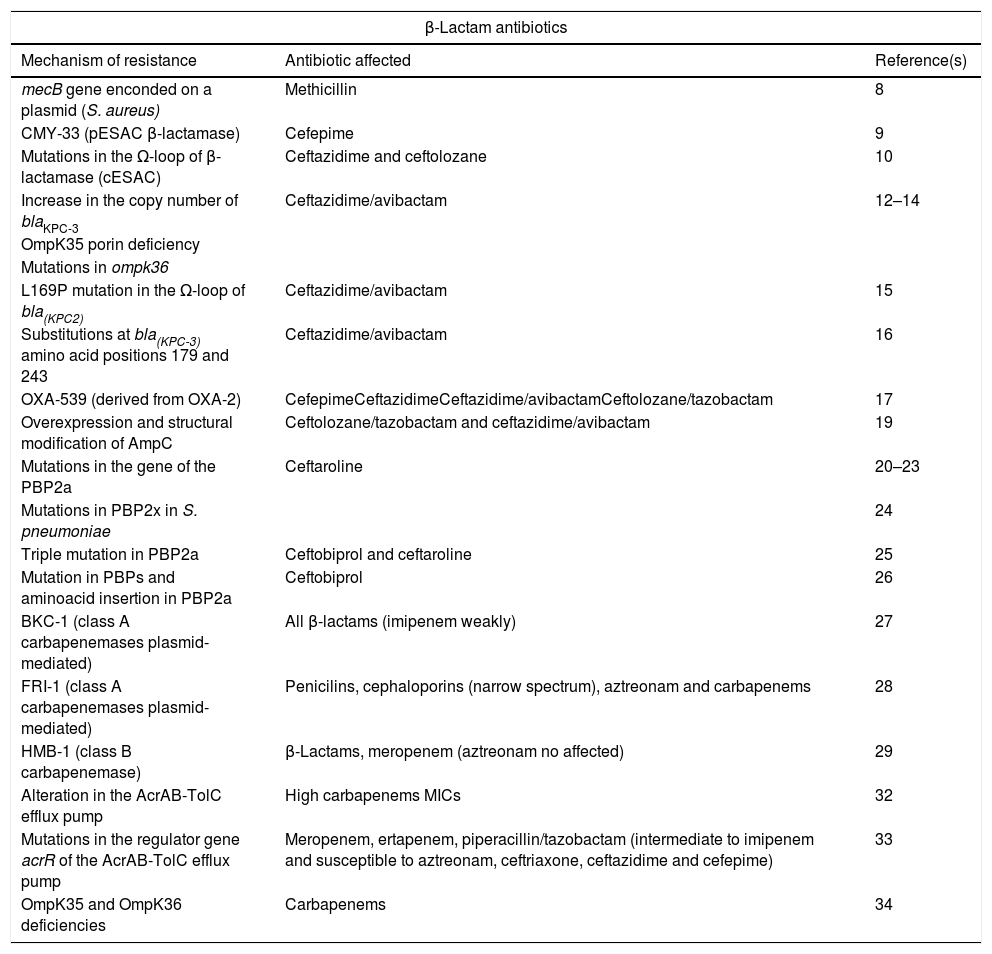
Summary of new mechanisms of resistance to beta-lactam antibiotics.
| β-Lactam antibiotics | ||
|---|---|---|
| Mechanism of resistance | Antibiotic affected | Reference(s) |
| mecB gene enconded on a plasmid (S. aureus) | Methicillin | 8 |
| CMY-33 (pESAC β-lactamase) | Cefepime | 9 |
| Mutations in the Ω-loop of β-lactamase (cESAC) | Ceftazidime and ceftolozane | 10 |
| Increase in the copy number of blaKPC-3 | Ceftazidime/avibactam | 12–14 |
| OmpK35 porin deficiency | ||
| Mutations in ompk36 | ||
| L169P mutation in the Ω-loop of bla(KPC2) | Ceftazidime/avibactam | 15 |
| Substitutions at bla(KPC-3) amino acid positions 179 and 243 | Ceftazidime/avibactam | 16 |
| OXA-539 (derived from OXA-2) | CefepimeCeftazidimeCeftazidime/avibactamCeftolozane/tazobactam | 17 |
| Overexpression and structural modification of AmpC | Ceftolozane/tazobactam and ceftazidime/avibactam | 19 |
| Mutations in the gene of the PBP2a | Ceftaroline | 20–23 |
| Mutations in PBP2x in S. pneumoniae | 24 | |
| Triple mutation in PBP2a | Ceftobiprol and ceftaroline | 25 |
| Mutation in PBPs and aminoacid insertion in PBP2a | Ceftobiprol | 26 |
| BKC-1 (class A carbapenemases plasmid-mediated) | All β-lactams (imipenem weakly) | 27 |
| FRI-1 (class A carbapenemases plasmid-mediated) | Penicilins, cephaloporins (narrow spectrum), aztreonam and carbapenems | 28 |
| HMB-1 (class B carbapenemase) | β-Lactams, meropenem (aztreonam no affected) | 29 |
| Alteration in the AcrAB-TolC efflux pump | High carbapenems MICs | 32 |
| Mutations in the regulator gene acrR of the AcrAB-TolC efflux pump | Meropenem, ertapenem, piperacillin/tazobactam (intermediate to imipenem and susceptible to aztreonam, ceftriaxone, ceftazidime and cefepime) | 33 |
| OmpK35 and OmpK36 deficiencies | Carbapenems | 34 |
pESAC: extended-spectrum AmpC plasmid-mediated; cESAC: extended-spectrum AmpC chromosomal.
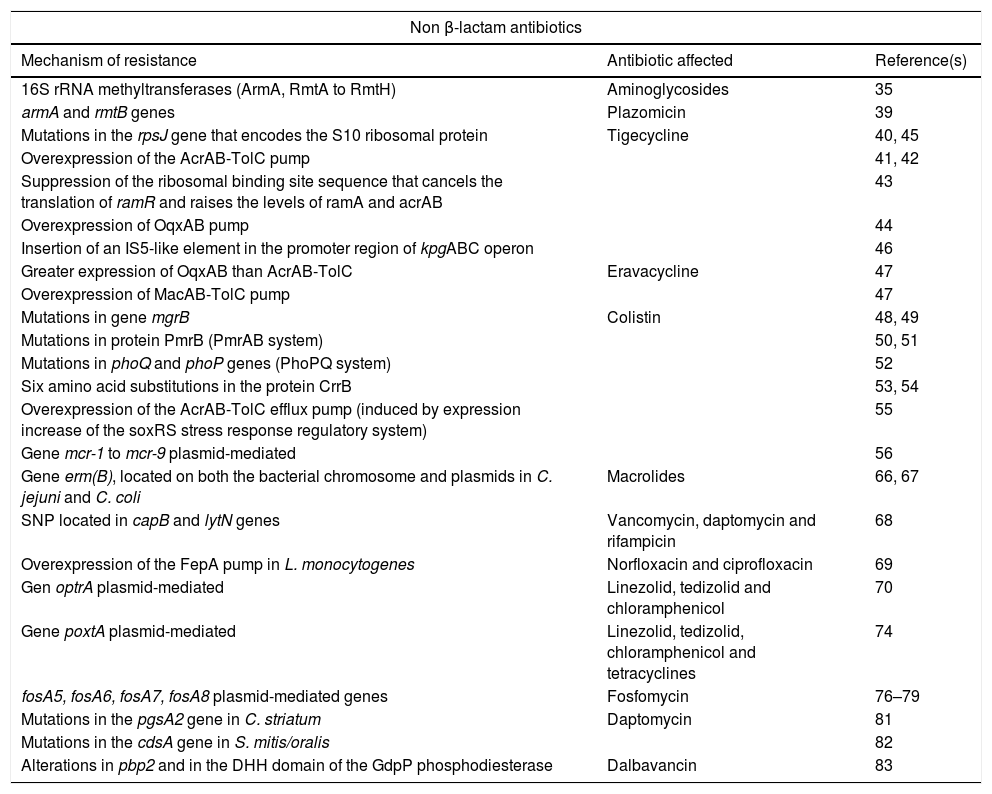
Summary of new mechanisms of resistance to non beta-lactam antibiotics.
| Non β-lactam antibiotics | ||
|---|---|---|
| Mechanism of resistance | Antibiotic affected | Reference(s) |
| 16S rRNA methyltransferases (ArmA, RmtA to RmtH) | Aminoglycosides | 35 |
| armA and rmtB genes | Plazomicin | 39 |
| Mutations in the rpsJ gene that encodes the S10 ribosomal protein | Tigecycline | 40, 45 |
| Overexpression of the AcrAB-TolC pump | 41, 42 | |
| Suppression of the ribosomal binding site sequence that cancels the translation of ramR and raises the levels of ramA and acrAB | 43 | |
| Overexpression of OqxAB pump | 44 | |
| Insertion of an IS5-like element in the promoter region of kpgABC operon | 46 | |
| Greater expression of OqxAB than AcrAB-TolC | Eravacycline | 47 |
| Overexpression of MacAB-TolC pump | 47 | |
| Mutations in gene mgrB | Colistin | 48, 49 |
| Mutations in protein PmrB (PmrAB system) | 50, 51 | |
| Mutations in phoQ and phoP genes (PhoPQ system) | 52 | |
| Six amino acid substitutions in the protein CrrB | 53, 54 | |
| Overexpression of the AcrAB-TolC efflux pump (induced by expression increase of the soxRS stress response regulatory system) | 55 | |
| Gene mcr-1 to mcr-9 plasmid-mediated | 56 | |
| Gene erm(B), located on both the bacterial chromosome and plasmids in C. jejuni and C. coli | Macrolides | 66, 67 |
| SNP located in capB and lytN genes | Vancomycin, daptomycin and rifampicin | 68 |
| Overexpression of the FepA pump in L. monocytogenes | Norfloxacin and ciprofloxacin | 69 |
| Gen optrA plasmid-mediated | Linezolid, tedizolid and chloramphenicol | 70 |
| Gene poxtA plasmid-mediated | Linezolid, tedizolid, chloramphenicol and tetracyclines | 74 |
| fosA5, fosA6, fosA7, fosA8 plasmid-mediated genes | Fosfomycin | 76–79 |
| Mutations in the pgsA2 gene in C. striatum | Daptomycin | 81 |
| Mutations in the cdsA gene in S. mitis/oralis | 82 | |
| Alterations in pbp2 and in the DHH domain of the GdpP phosphodiesterase | Dalbavancin | 83 |
Grampositive microorganisms with resistance to multiple antibiotics are responsible for both infections associated with hospital and community care. Methicillin-resistant Staphylococcus aureus (MRSA), glycopeptide-resistant Enterococcus faecium (GRE) and Streptococcus pneumoniae with decreased penicillin susceptibility are considered a public health problem according to the CDC.4 In addition, the first two are classified as “High Priority” pathogens in the list of priority bacteria established by WHO for the development of new antibiotics.5 Although MRSA and GRE infections have traditionally been associated with the hospital environment, cases of MRSA and linezolid-resistant enterococci infections acquired in the community are becoming more frequent.
Despite the clinical and epidemiological importance of antibiotic resistance in grampositive microorganisms, the greatest impact is due to the strains of gramnegative bacteria MDR and XDR. In fact, WHO considers enterobacteria, Acinetobacter baumannii and Pseudomonas aeruginosa resistant to carbapenems antibiotics, pathogens of “critical priority” for the development of new antibiotics.6
Resistance to β-lactam antibioticsMethicillinThe treatment of S. aureus is complicated due to the increase in the resistance rates, especially against β-lactam antibiotics. This is a consequence of the acquisition of the mecA or mecC genes that encode penicillin-binding protein 2a (PBP2a) or PBP 2c, respectively, that determine the expression of resistance to oxacillin and, consequently, to almost all β-lactams. These genes are located in the staphylococcal chromosomal cassette (SCCmec) of which several types have been described, some of which contain, in addition to the mecA gene, other genes that encode resistance to different non-β-lactam antimicrobials and are associated, in general, with hospital strains.7
In 2018, during a routine MRSA screening, it was discovered a S. aureus isolate carrying a mecB gene encoded on a plasmid, that was previously described in Macrococcus caseolyticus but not in any staphylococcal species. This plasmid-encoded, and thereby transferable, methicillin resistance mechanism reveals a novel level risk of the transfer of broad β-lactam resistance in staphylococci.8 Further studies are needed to clarify the real prevalence of mecB-caused methicillin resistance among MRSA and methicillin-resistant coagulase-negative staphylococci in humans.8
CephalosporinsCephalosporins and cephalosporins with β-lactamase inhibitorsEnterobacteria may be resistant to third-generation cephalosporins, in many cases without affecting the carbapenems, as a consequence of the production of extended-spectrum β-lactamases (ESBL), AmpC type β-lactamases, both chromosomal (cAmpC) and plasmid-mediated (pAmpC), and by the sum of mechanisms including the decrease of permeability and efflux pumps. Sporadically, cases of strains carrying variants of cAmpC with the ability to hydrolyze cefepime, known as extended spectrum cAmpC (cESAC), have been reported.
ESAC mediated by plasmids (pESAC) have also been described. In 2015, Pires et al.9 isolated a clinical strain of E. coli producing pAmpC CMY-2 susceptible to cefepime which, after treatment with this drug, suffered a double deletion of amino acids (Leu293-Ala294) in the H-10 helix of the enzyme. The new pESAC β-lactamase originated, CMY-33, had a phenotype similar to an ESBL (high cefepime MIC and a lower cefoxitin MIC).9
In Pseudomonas aeruginosa, the cESAC have been less studied due to the high polymorphism in the cAmpC sequence of this species. A study conducted by Berrazeg et al.10 showed that the presence of these variants, resulting from mutations in the Ω-loop of the β-lactamase (substrate binding site that represents a “hot spot” for mutations that favor catalytic activity and extend the spectrum of β-lactamase activity), have therapeutic implications as they result in conformational changes that increase the resistance to ceftazidime and ceftolozane. The MICs of cefepime and aztreonam were 16mg/L in this case.10
Recently, some new antibiotics or combinations of antibiotics with β-lactamase inhibitors, such as ceftolozane/tazobactam, ceftazidime/avibactam and meropenem/vaborbactam, have contributed to partially alleviating the lack of therapeutic alternatives against gramnegative resistant pathogens. In addition, other drugs with activity against these pathogens are currently in the process of marketing authorization (imipenem/relebactam, aztreonam/avibactam, and cefiderocol).
In 2014, the Food and Drug Administration approved the combination of ceftazidime/avibactam, which is active mainly against gramnegative bacteria producing carbapenemases of the Amber class A (such as KPC) and OXA-48-like (class D), and against AmpC and ESBL. Simultaneously ceftolozane/tazobactam was approved, whose main contribution is the activity against MDR P. aeruginosa given its stability against the cAmpC of this species. However, quite soon resistance to these new drugs emerged, and although the percentages of resistance to date are low, this fact is a cause of concern.
In 2015, the first case of ceftazidime/avibactam resistance in a clinical strain of a KPC-producing Klebsiella pneumoniae was published.11 Since then, different studies have been carried out to clarify the mechanism of resistance against this new drug. It is currently known that the increase in the copy number of blaKPC-3 in the presence of an ESBL-carrying plasmid, and the alteration in the permeability due to the OmpK35 porin deficiency and/or the presence of mutations in ompk36, contribute to resistance to ceftazidime/avibactam.12–14 The overexpression of blaKPC-3 is due to the transposition of the transposon carrying the gene to a second plasmid carrying ESBL genes, resulting in an increase in the number of copies of blaKPC-3 and in the hydrolysis of ceftazidime.13
In addition, cases of resistance to ceftazidime/avibactam among K. pneumoniae isolates during the course of treatment have also been described. They have been associated with mutations in the blaKPC gene.15,16 In strains carrying KPC-2-like carbapenemases, the mutations have been located in the Ω-loop, the active site of class A β-lactamases,15 while in those with KPC-3-like carbapenemases, the most frequent variations have been detected in the amino acid Ambler positions 179 (tyrosine by aspartic acid) and 243 (methionine by threonine).16 In some of the isolates with mutations in blaKPC-3, the recovery of susceptibility to meropenem is remarkable, as well as a decrease of cefepime and ceftriaxone MICs, although without restoring the susceptibility to the latter. The efficacy of meropenem in the treatment of these isolates is unidentified, as the phenotypic stability of this phenomenon in the clinic is unknown.16
In 2017, Fraile-Ribot et al.17 described a new OXA-type extended-spectrum β-lactamase, OXA-539, derived from OXA-2, on which a duplication of a key residue of aspartate had been observed. Through transformation studies in the PAO1 strain, the MICs of cefepime, ceftazidime, ceftazidime/avibactam and ceftolozane/tazobactam against the transformant carrying OXA-539 were higher than these against isolates carrying OXA-2, the opposite that happened with meropenem MIC.17
Published reports regarding the emergence of resistance in P. aeruginosa after ceftolozane/tazobactam administration, are limited.17,18 Whole-genome sequencing studies conclude that the development of resistant strains requires the accumulation of several alterations including, at least, those that lead to the overexpression and structural modification of AmpC, as amino acid mutations or deletions.19 It has been observed that those strains that develop resistance to ceftolozane/tazobactam show cross resistance to ceftazidime/avibactam, however, the MICs of piperacillin/tazobactam and carbapenems decreased slightly.19
Ceftaroline and ceftobiproleIn order to address β-lactam resistance in grampositive bacteria, two so-called fifth-generation cephalosporins, ceftaroline and ceftobiprole, have recently been marketed, whose mechanism of action stands out above others for its high affinity for PBP2a.
However, ceftaroline resistant strains have already been described, being the emergence of mutations in the gene that encodes the PBP2a the main mechanism of resistance. These mutations are single nucleotide polymorphisms (SNP) that cause an amino acid change, altering the electrostatic potential and preventing the binding of the ceftaroline molecule.20 Such change can be located either directly in the penicillin binding pocket in the transpeptidase region21 or in the allosteric domain of the protein.22 The first would be responsible for high level resistance while the latter is associated with a lower resistance level.23
Regarding Streptococcus pneumoniae, a clinical strain resistant to ceftaroline has recently been described in which mutations in several PBPs were detected, including the PBP2x. This PBP has shown the greatest affinity for ceftaroline and therefore mutations located in it are associated with resistance to this new cephalosporin.24
Concerning ceftobiprole, there are few studies aimed at determining the mechanism of resistance against this antimicrobial, most of them performed in laboratory mutants and not in clinical isolates. A triple mutation has been described in PBP2a (N146K-N204K-G246E) responsible for causing high resistance to ceftaroline and ceftobiprole in African strains of MRSA.25 Morroni et al.26 analyzed the PBP of MRSA strains resistant to this antibiotic isolated in clinical samples and observed amino acid substitutions in all investigated PBP (from PBP1 to PBP4) and a new 5 amino acid insertion in PBP2a that could be involved in resistance to this new cephalosporin without affecting resistance to other β-lactams.26
The emergence of resistance against these new antibiotics results in the loss of the only β-lactams clinically active against MRSA.
Carbapenemem antibioticsIn general, the main mechanisms of resistance to carbapenems in gramnegative bacteria are: (i) the expression of enzymes with hydrolytic capacity (carbapenemases), and (ii) the overexpression of β-lactamases (ESBL and cephalosporinases) without or with little activity against this class of antibiotics in combination with a decrease in permeability due to modifications in porins.
The carbapenemases clinically relevant are classified in class A, B and D. Until now, the known class A types were NmcA/IMI, SME, KPC and GES with their different variants. In recent years, two new types of class A plasmid-mediated carbapenemases have been described: BKC-1 in a Brazilian strain of K. pneumoniae,27 and FRI-1 in a strain of Enterobacter cloacae.28 BKC-1 has a weak ability to hydrolyze imipenem, but is responsible for producing resistance to all tested β-lactams (penicillins, cephalosporins, monobactams, and carbapenems).27 However, FRI-1 confers high resistance to penicillins, narrow spectrum cephalosporins, aztreonam and carbapenems without conferring significant resistance to broad spectrum cephalosporins (ceftazidime, cefotaxime and cefepime).28
Class B carbapenemases belong mainly to three types NDM, VIM and IMP, of which several variants have been described. In 2017, a strain of P. aeruginosa producing a new class B carbapenemase, HMB-1,29 was isolated in Germany. In that strain, the blaHMB-1 gene was detected on the chromosome and the genetic homology with the closest gene, blaKHM-1 was 74.3%. In transformation studies, the MIC of meropenem was increased in six double dilutions in an E. coli receptor, but its aztreonam susceptibility did not vary.29
On the other hand, gramnegative bacteria can acquire resistance, generally of low level, to different families of antibiotics by the presence of efflux pumps. In particular, the overexpression of the AcrAB-TolC efflux pump, belonging to the RND family (resistance-nodulation-cell division), confers clinically relevant resistance to various antibiotics used in the treatment of infections caused by these bacteria.30,31 Its importance concerning the resistance to carbapenems has been poorly studied to date but its clinical implication has recently been observed. A study with laboratory mutants showed that carbapenemase MICs in carbapenemase-producing enterobacteria strains with alterations in AcrAB-TolC were higher in comparison with strains without efflux-pump alterations.32 The report of a clinical isolate of Klebsiella oxytoca with an unusual phenotype has recently been published: resistant to ertapenem, meropenem and piperacillin-tazobactam, intermediate susceptibility to imipenem, and susceptible to aztreonam and several third and fourth generation cephalosporins (ceftriaxone, ceftazidime and cefepime).33 After sequencing the genome, mutations in the regulator gene acrR were detected. These changes conditioned an overexpression of this efflux pump, as well as deficiencies in the porins OmpK36 and OmpK35,33 previously associated with carbapenem resistance.34
Resistance to non-β-lactam antibioticsAminoglycosidesThe worldwide emergence of 16S rRNA methyltransferases is a growing concern due to their ability to confer high-level resistance to all clinically relevant aminoglycosides. Although most of them were described for the first time before 2014, the start date of this review, below are some important subsequent milestones in relation to its dissemination. A total of 9 different 16S rRNA methyltransferases have been described, including ArmA, and from RmtA to RmtH.35 Some of them have different variants such as the new RmtB4 recently described.36 Recent studies show that these methyltransferases are spreading mainly among carbapenemase-producing Enterobacteriaceae.35,37 This association poses a risk for the treatment of multidrug-resistant gramnegative infections in the clinical setting. The most frequently described 16S rRNA methyltransferases are ArmA and RmtB, RmtC, and RmtF, in general associated to NDM or KPC-carbapenemase-producing isolates.35–39
PlazomicinThis antimicrobial is a next-generation aminoglycoside developed to overcome the common aminoglycoside-resistance mechanisms (aminoglycoside-inactivating enzymes) for the treatment of patients with serious infections caused by multidrug-resistant Enterobacteriaceae, including ESBL-producing and carbapenem-resistant. In a recent study carried out in Greek hospitals,39 plazomicin retained activity against most carbapenemase-producing K. pneumoniae, with MICs consistently lower than those of other aminoglycosides, even in the presence of aminoglycoside modifying enzymes; however, 7.7% of isolates (16 KPC, 6 VIM, and one KPC plus OXA-48-producers) were resistant to plazomicin and harbored rmtB (n=22) or armA (n=1) genes.39
TetracyclinesTigecyclineTigecycline is a derivative of minocycline, belonging to the glycylcyclines family, which has activity against a broad spectrum of pathogens.
There are few studies of tigecycline resistance in grampositive microorganisms, but some of these, carried out in selected mutants in vitro, have shown that resistance to this antibiotic in S. aureus is related to the expression of the MepA efflux pump.
However, the mechanisms of resistance to tigecycline in enterococci were unknown until 2015, when Niebel et al.40 detected the association of tigecycline resistance in clinical strains of E. faecium with the presence of mutations in the rpsJ gene, which encodes the S10 structural protein of the 30S ribosomal subunit.40
Regarding gramnegative bacteria, tigecycline is currently one of the few options available for the treatment of infections caused by MDR and XDR microorganisms. Unfortunately, the emergence of resistance against this molecule is increasing.
The best known resistance mechanism is the overexpression of the AcrAB-TolC pump due to mutations in the coding region of regulatory genes.41 This mechanism has been studied primarily in K. pneumoniae but has also been observed in strains of other enterobacteria such as E. cloacae.42 Ye et al.43 track clinical strains of K. pneumoniae in order to study the in vivo development of resistance to tigecycline; they detected its rapid emergence after 41 days of treatment due to a suppression of the ribosomal binding site sequence that cancels the translation of ramR and raises the levels of ramA and acrAB.43 On the other hand, overexpression of the OqxAB pump has also been associated with resistance to tigecycline.44
Recent studies have shown that resistance to this antibiotic can occur by mechanisms independent of the AcrAB system. In 2014, an isolate of KPC-3-carbapenemase producing K. pneumoniae belonging to the high-risk clone ST512 and resistant to tigecycline but without ramR alterations was reported; in this case the resistance was due to an amino acid change consequence of a mutation in the rpsJ gene that encodes the S10 ribosomal protein.45 This protein belongs to the 30S subunit of ribosomal RNA and is the binding target of this drug.45 During the same year, the increase in the expression of the kpgABC operon was discovered in a resistant strain, due to the insertion of an IS5-like element in the promoter region, which was linked to the reduction of tigecycline susceptibility in vivo in a K. pneumoniae isolate.46
EravacyclineEravacycline is the first fluorocycline, derived from minocycline, with activity against aerobic and anaerobic grampositive and gramnegative bacteria. In resistant and heteroresistant strains, a greater expression of OqxAB than AcrAB-TolC has been observed, although overexpression of the ramA regulatory gene is remarkable without the consequent alteration of the AcrAB-TolC pump. It has been seen that the MacAB-TolC pump also plays an important role in the development of resistance to this new drug.47
ColistinThe overall increase in carbapenemase-producing enterobacteria has led to an increase in the use of colistin, an old antibiotic with a high toxicity potential which however, has become one of the few therapeutic alternatives to infections due to some gramnegative XDR bacteria. New mechanisms of resistance to this family of antibiotics (polymyxins) both chromosomal and plasmid mediated, have been described in recent years. Chromosomal mechanisms confer acquired resistance to polymyxins mediated by the addition of cationic groups (L-Ara4n and pEtN) to lipopolysaccharide lipid A (LPS). Sugar synthesis requires the products of the pmrE gene and the pmrHFIJKLM operon; the systhesis of pEtN is encoded by the pmrC gene. These modifications generate a positively charged LPS and this reduces its affinity for polymyxins, also positively charged. The overexpression of transcriptional regulatory systems that control LPS modifications is due to mutations and is the most common chromosomal resistance mechanism. In K. pneumoniae, alterations in mgrB,48,49 in PmrB (PmrAB system), selected in vivo after treatment with low dose of colistin50,51 and in phoQ and phoP genes (PhoPQ system), have been identified.52
Another regulatory system that has been related to colistin resistance is CrrAB. Six amino acid substitutions in the protein CrrB have been identified as responsible for this resistance. This new mechanism is dependent on the genome of the bacterium since the gene is not present in all strains of K. pneumoniae and is absent in E. coli.53,54
In K. pneumoniae strains heteroresistant to colistin, the presence of a partial nucleotide deletion in the phoP gene has been related to the reversion to a phenotype susceptible to this antibiotic.53 However, this mechanism is not involved in the emergence of heteroresistance in strains of Enterobacter asburiae in which it has been associated with an expression increase of the soxRS stress response regulation system, which induces overexpression of the AcrAB-TolC efflux pump.55
By the end of 2015, resistance to colistin was thought to occur exclusively via chromosomal mutations, but that year the emergence in China of a horizontally and plasmid-mediated transmissible mechanism known as mcr-156 was reported. This gene encodes a phosphoethanolamine transferase that modifies lipid A by adding phosphoethanolamine. In bacteria carrying this gene, the MIC of polymyxin increases from 4 to 8-fold, so its acquisition is sufficient to confer resistance to colistin in different enterobacteria.56
Since this critical finding, the gene has been actively searched in existing collections of strains, and isolates carrying this gene have been documented from dates prior to its description.57–59 In human strains, transferable resistance to colistin has been identified mainly in isolates of patients who had not previously been treated with colistin and its acquisition has been associated with ingestion of contaminated food resulting in an intestinal colonization.59 Co-production of mcr-1 and other resistance mechanisms such as ESBL60 or carbapenemases such as NDM-5,61 VIM-162 or KPC63 has been described, among others.
To date, it has been described in enterobacteria the presence of mcr-1 to mcr-9 genes showing high genetic variations.64,65 However, variants within these with nucleotide variations have been described in genes belonging to the same group (mcr-1.1, mcr-1.2).63
MacrolidesMacrolide resistance is mainly due to three mechanisms: (i) modification of the target by mutations or methylation, (ii) inactivation of the antibiotic, and (iii) active efflux pump. Ribosomal methylation erm gene-mediated is a mechanism that confers resistance to macrolides and acquires great relevance in some grampositive species.
Historically, a low incidence of resistance to macrolides has been reported in Campylobacter spp., mainly due to two mechanisms: (i) target mutations (23S rRNA or in the rplD and rplV genes encoding the L4 and L22 ribosomal proteins) and (ii) efflux pumps (CmeABC). Recently, the presence of the erm(B) gene, located on both the bacterial chromosome and plasmids,66,67 has been identified for the first time in Campylobacter jejuni and Campylobacter coli strains.
VancomycinVancomycin is a glycopeptide antibiotic approved in 1958 that continues to be used for the treatment of infections due to grampositive microorganisms.
The mechanisms of glycopeptide resistance in S. aureus and enterococci are well known, especially the increase in the thickness of the bacterial wall (S. aureus) and the acquisition of the vanA and vanB genes (enterococci). Since its introduction in clinical practice, S. aureus strains with intermediate susceptibility (VISA) and with high vancomycin resistance (VRSA), have been described, although the isolation of the latter remains very rare. However, the impact of glycopeptide resistance among the genus Enterococcus is greater.
It was in 2015 when Yamaguchi et al.68 isolated a clinical strain of MRSA resistant to vancomycin. After carrying out sequencing studies, they detected SNP located in two genes until then not related to vancomycin resistance: capB, which encodes a tyrosine kinase involved in the biosynthesis of the capsular polysaccharide, and lytN, which encodes a peptidoglycan hydrolase called N-acetylmuramil-l-alaninaamidase which participates in bacterial growth. This strain also showed resistance to daptomycin and rifampicin. The authors concluded that the increasing vancomycin MIC, in the absence of van genes, was due to these new substitutions.68
FluoroquinolonesAlthough resistance to fluoroquinolones in grampositive bacteria of clinical interest is mainly due to mutations in the parC and gyrA chromosomal genes, in othe grampositives like Listeria monocytogenes it had previously been linked to efflux pumps of the MFS (major superfamily facilitator) family.
Recently, resistance to norfloxacin and ciprofloxacin in this species has been associated with overexpression of the FepA pump, belonging to the MATE (multidrug and toxic compound extrusion) family.69
LinezolidLinezolid was the first oxazolidinone introduced in clinical practice and is increasingly used for the treatment of infections due to MRSA and enterococci resistant to glycopeptides. Until 2015, the most frequent linezolid resistance mechanism in enterococci was chromosomal mutations in the subunit 23S of ribosomal RNA, mainly the G2576T change.
That year a new transferable gene, optrA, was described for the first time in Enterococus faecalis and Enterococcus faecium isolated in China; this gene is able to confer resistance to both the oxazolidinone group (linezolid and tedizolid) and the phenicol group (chloramphenicol).70 Since its description, the optrA gene has been linked to enterococci, although it has also been described in strains of Staphylococcus suis of porcine origin. The fact that identical variants of optrA, present in strains unrelated to each other, have been detected in presumably identical plasmids indicates a spread of the plasmid containing this gene.71 Its presence has been related to certain clones of E. faecalis such as ST480 and ST585.72,73
Later, the poxtA gene was described in the genome of a MRSA strain of clinical origin, responsible for resistance to oxazolidinones, phenicols and also to tetracyclines.74 Recently, this gene has been detected in a clinical strain of E. faecium,75 which would be explained for its location in a plasmid that confers the ability of interspecies transfer.
Both the oprtA gene and the poxtA gene encode proteins of the ATP-binding cassette F (ABC-F) family whose function is to modify the bacterial ribosome and thus protect it against the activity of such antibiotics.
The emergence of new genes transferable by plasmids warns of a possible spread not only among strains of the same bacterial species but also between different species that share the same ecological niche.
FosfomycinFosfomycin is a broad-spectrum cell wall synthesis inhibitor antibiotic. Production of chromosome or plasmid-encoded fosfomycin-inactivating enzymes Fos is a mechanism of resistance known since many years ago. The FosA genes are mostly found in Enterobacterales, Pseudomonas spp. and Acinetobacter spp. But new gene subtypes plasmid-mediated with similar structure have been identified in recent years: fosA5,76fosA6,77fosA778 and fosA8.79 Concurrence in plasmids of these genes with other genes conferring resistance to beta-lactams, aminoglycosides and fluoroquinolones has also been described.77
DaptomycinThis lipopeptide has bactericidal activity against a wide range of grampositive pathogens and has become a widely used option in the management of S. aureus infections. Daptomycin interacts with the bacterial cell membrane and is dependent of phosphatidylglycerol (PG) concentration.
It is known that resistance to daptomycin in S. aureus is mainly due to mutations in the mprF gene encoding the bifunctional enzyme MprF, responsible for incorporating a positive charge into the bacterial peptidoglycan. These mutations increase this function causing an electrostatic repulsion of the daptomycin-Ca2+ complex and preventing its binding to the target.
In addition, mutations in yycG (gene encoding a histidine kinase) and in rpoB and rpoC (genes encoding subunits of RNA polymerase) have been detected in S. aureus strains with a daptomycin MIC higher than the susceptible range. Daptomycin resistance in enterococci appears to be mediated by mechanisms not yet well-established but different from those detected in S. aureus.80
In recent years, daptomycin resistance has emerged in other clinically relevant species such as Corynebacterium striatum or Streptococcus mitis. The mechanism described in a strain of C. striatum with high resistance to daptomycin, acquired during treatment, is based on mutations in the pgsA2 gene encoding a peptidoglycan synthetase and generating a significant loss of peptidoglycan. It seems that these mutations are sufficient to determine a high level of resistance.81
S. mitis/oralis appears to be less susceptible to daptomycin than other streptococci belonging to the viridans group and tends to develop high resistance rapidly. It has been observed that mutations in the cdsA gene, which encodes a phosphatidate citidyltransferase, entail the loss of function of this enzyme involved in the synthesis of cell membrane phospholipids and lead to the absence of peptidoglycan and cardiolipin, which results in a daptomycin resistance increase.82
DalbavancinTo date, very few strains with acquired resistance during treatment with this new lipoglycopeptide have been described, but recently small colony variants of MRSA, with decreased susceptibility to dalbavancin and resistance to teicoplanin, have been isolated from a patient.83 In this study, an increase in wall thickness was observed, as well as a damaged separation of cells with multiple or incomplete cross-links, which translates into significantly reduced bacterial growth rates. After sequencing, eight alterations were detected in different genome sequences, being those located in pbp2 and in the DHH domain of the GdpP phosphodiesterase, those most likely related to the observed cellular alteration.83 An SNP in the pbp2 gene could interfere with the structure of the PBP2 protein by altering its activity, and the 534bp deletion found in the DHH domain of the GdpP could contribute to the alteration of its enzymatic activity.83
Future perspectivesThe progress of bacterial resistance to antibiotics is inevitable and in a globalized world it is difficult to control.
New ways of research are needed, mainly focused on the development of new antibiotics with different mechanisms of action than those known to date. Another interesting line of research is the study of the collaboration between the human immune system and the antibiotics, as well as the role of the commensal microbiota in the control and eradication of invasive bacteria. The success of fecal transplantation, used in recurrent Clostridium difficile infections, could be adapted and be a possible developmental path to follow.
Currently, and given the delay in the appearance of new antibiotics, the use of those available should be optimized without forgetting old drugs such as colistin or fosfomycin, which have proven to be a good therapeutic option in the fight against multidrug-resistant bacteria.
Conflict of interestThe authors declare that they have no conflict of interest.
This study was supported by Fundación Soria Melguizo. The funders had no role in study design or the decision to submit the work for publication.
The authors thank Nora Ramos for helping with the English translation.