Most living beings are able to perform actions that can be considered intelligent or, at the very least, the result of an appropriate reaction to changing circumstances in their environment. However, the intelligence or intellectual processes of humans are vastly superior to those achieved by all other species. The adult human brain is a highly complex organ weighing approximately 1500g, which accounts for only 2% of the total body weight but consumes an amount of energy equal to that required by all skeletal muscle at rest. Although the human brain displays a typical primate structure, it can be identified by its specific distinguishing features.
The process of evolution and humanisation of the Homo sapiens brain resulted in a unique and distinct organ with the largest relative volume of any animal species. It also permitted structural reorganisation of tissues and circuits in specific segments and regions. These steps explain the remarkable cognitive abilities of modern humans compared not only with other species in our genus, but also with older members of our own species.
Brain evolution required the coexistence of two adaptation mechanisms. The first involves genetic changes that occur at the species level, and the second occurs at the individual level and involves changes in chromatin organisation or epigenetic changes. The genetic mechanisms include: (a) genetic changes in coding regions that lead to changes in the sequence and activity of existing proteins; (b) duplication and deletion of previously existing genes; (c) changes in gene expression through changes in the regulatory sequences of different genes; and (d) synthesis of non-coding RNAs.
Lastly, this review describes some of the main documented chromosomal differences between humans and great apes. These differences have also contributed to the evolution and humanisation process of the H. sapiens brain.
La mayor parte de los seres vivos son capaces de realizar acciones que pueden ser consideradas inteligentes o al menos el resultado de un proceso de reacción adecuado ante las circunstancias cambiantes de su medio ambiente. Sin embargo, la inteligencia o los procesos intelectuales que desarrollan los seres humanos son enormemente superiores a los que logran los organismos de cualquier otra especie. El cerebro humano adulto es un órgano sumamente complejo: pesa aproximadamente 1.500g, lo que representa solo el 2% del peso corporal pero consume igual cantidad de energía que todo el músculo esquelético en reposo. Aunque el cerebro humano presenta una estructura típicamente primate, revela algunas características que lo distinguen y lo individualizan plenamente.
El proceso de evolución y humanización del cerebro del Homo sapiens (H. sapiens) lo convirtió en un órgano único y diferente, alcanzando el mayor tamaño relativo entre todas las especies, pero además le permitió una reorganización estructural de tejidos y circuitos en segmentos y regiones específicas. Esto explica las notables capacidades cognitivas del hombre moderno, en comparación no solo con otros miembros de su género, sino con otros miembros más antiguos de su propia especie.
La evolución del cerebro requirió la coexistencia de 2 mecanismos de adaptación. El primero involucra cambios genéticos que ocurren a nivel de especies y el segundo ocurre a nivel individual e involucra cambios en la organización de la cromatina o cambios epigenéticos. Entre los mecanismos genéticos se encuentran: a) cambios en regiones genéticas codificantes que conducen a cambios en la secuencia y actividad de proteínas existentes; b) los procesos de duplicación y deleción de genes previamente existentes; c) cambios en la expresión génica a través de modificaciones en las secuencias reguladoras de diferentes genes, y d) síntesis de ARNs no codificantes.
Finalmente, en esta revisión se describen algunas de las más importantes diferencias cromosómicas reportadas entre humanos y grandes simios, que también han contribuido al proceso de evolución y humanización del cerebro del H. sapiens.
How intelligence should be defined continues to be subject to debate. However, it is widely accepted that the majority of animals, particularly those further up the evolutionary scale, perform a series of actions that may be considered intelligent, or at least the product of an appropriate reaction to a given set of environmental circumstances. In any case, human intelligence and intellectual processes are vastly different from those observed in any other species on the planet.
We know now that the brain is responsible for this capability in humans, although this has not always been the case: the ancient Egyptians extracted the brains from corpses during the process of mummification, while the ancient Greeks believed the brain to be no more than a radiator for cooling the blood flowing from the heart. In his magnum opus On the origin of species,1 Darwin's observations and discussions make little mention of the brain. Likewise, Darwin's contemporary Huxley2 argued that the human brain fundamentally resembled that of the great apes.3
The brain of an adult human is an intricate, complex organ weighing approximately 1500g and representing 2% of total body weight; however, it consumes the same amount of energy as all the body's skeletal muscle at rest.4 In general, the human brain is structured similar to a typical primate brain, but with certain distinguishing characteristics.
Despite the enormous body of biological and medical knowledge about the brain, there remain questions which are yet to be completely resolved. What makes the human brain so exceptional, compared even to the brains of the higher primates and ancestral hominids? When did our brain truly become human? How did the human brain evolve?
Initial attempts to answer these questions compared the human brain to those of species with genetic or evolutionary similarities (whether macroscopic or microscopic) in order to identify the similarities and differences. Attempts have also been made to compare the human brain to those of other members of the genus Homo, although a detailed comparison is impossible since the only remnants of our ancestors’ brains are prints left by contact with the endocranium.5 This has enabled us to understand the process of encephalisation that led to the development of the complex brain of Homo sapiens.
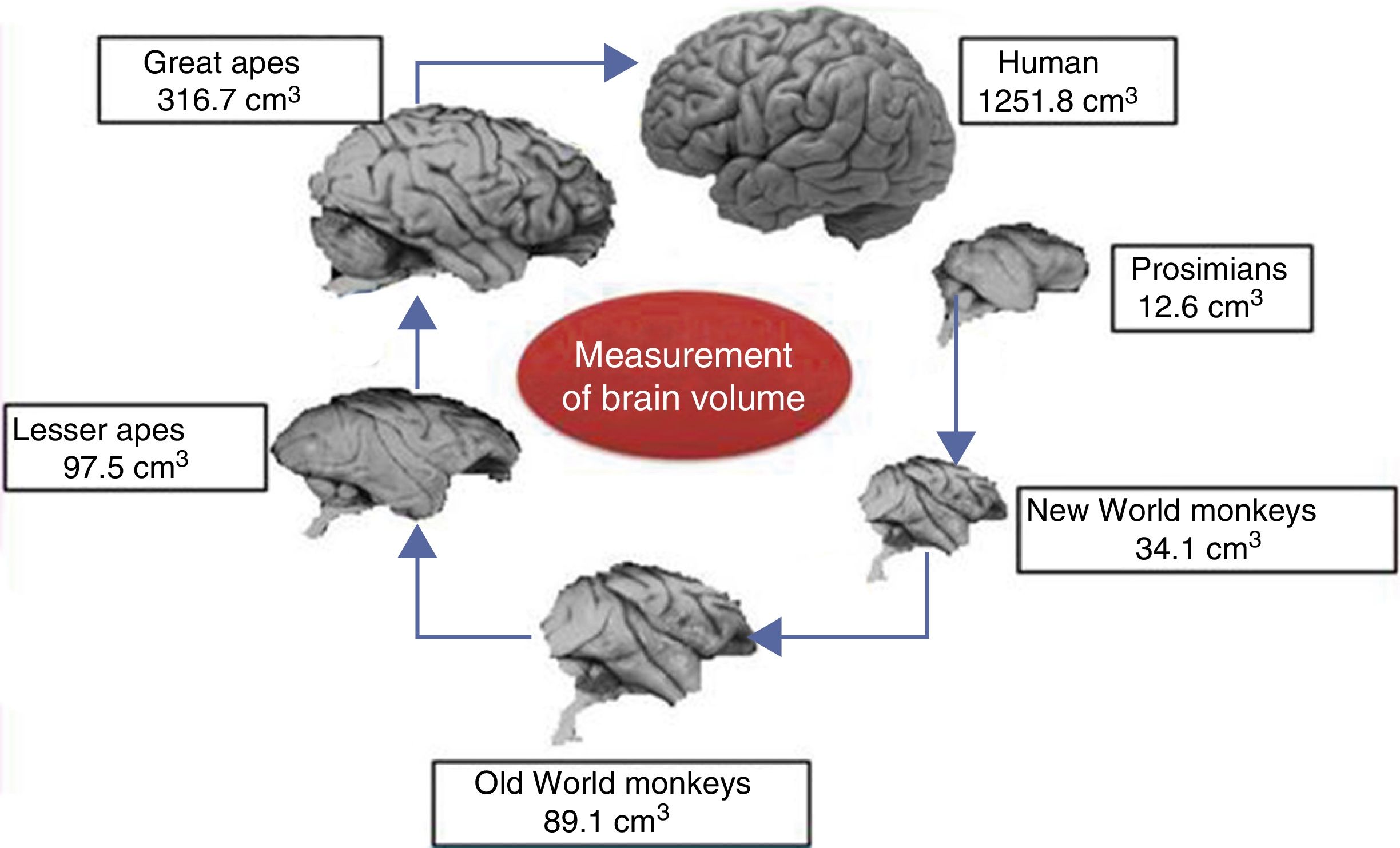
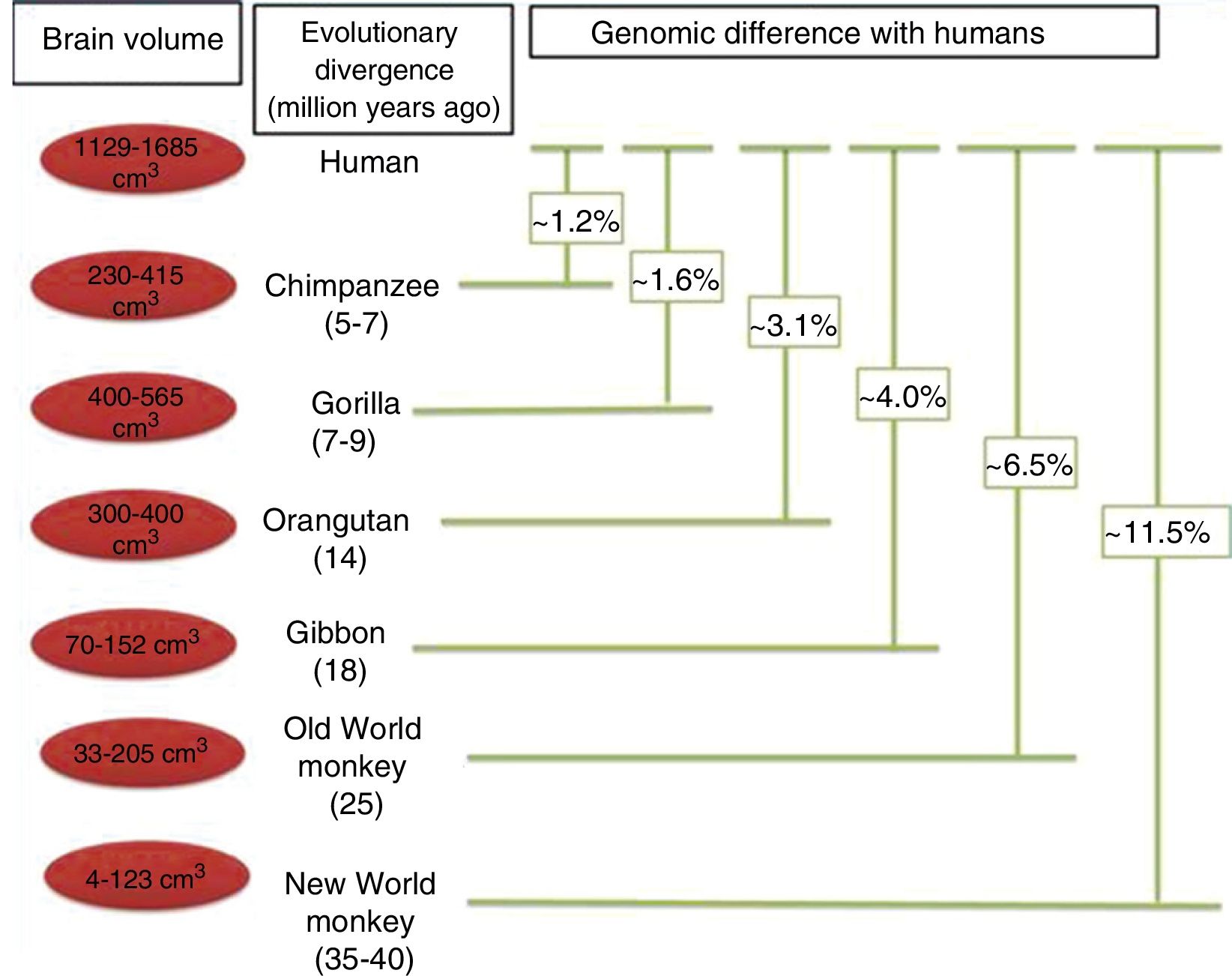
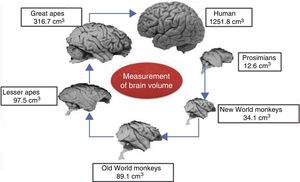
Brain size and the process of encephalisationThe human brain is large, with adult brains typically having a volume of approximately 1350cm3, weighing 1500g, and containing nearly 20billion neurons.6 It is therefore much larger than the brain of any extinct primate, and weighs 3 times more than the brain of the chimpanzee, our closest evolutionary relative (Fig. 1).7
A clear relation can be seen between total brain size and the attributes of a particular species. Brain size is a general predictor of mental capacity in non-human primates. Intelligence testing measures a species’ cognitive power according to the decisions taken by individuals in response to changing conditions or circumstances in a controlled environment.8 For practical ends, we can consider mental flexibility a type of intelligence. In prosimians and the great apes, cognitive flexibility increases in line with brain size. The small brain of the australopiths evolved in the same way into larger brains such as those of H. habilis and H. erectus, and ultimately into the large brain of H. sapiens.
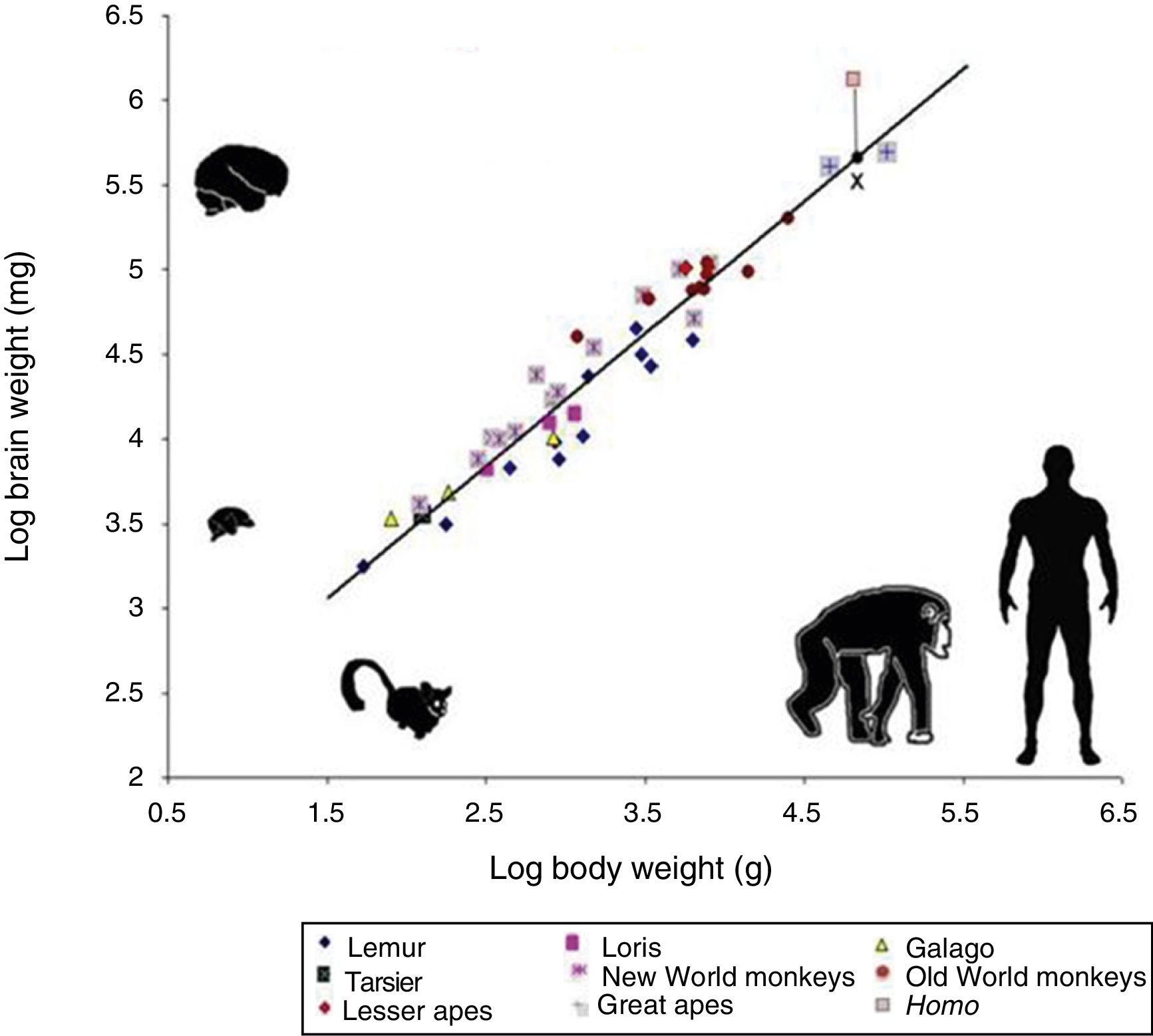
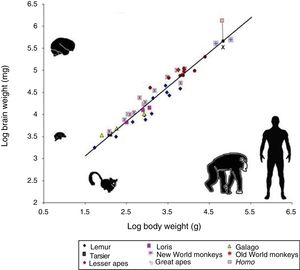
However, cognitive ability is not fully explained by total brain size. If this were the case, elephants (whose brain weighs 5kg) or whales (7.8kg) would be far more encephalised than humans, whose brain weighs only 1.5kg. Therefore, if we wish to attribute the uniqueness of the human brain to encephalisation, it should be measured in relative terms: brain weight as a proportion of total body weight (Fig. 2). However, this proportion is not an exact measure: for example, it would imply that shrews, whose brains weigh 0.25g, would be much more encephalised.
Logarithmic scale comparing total body weight (g) and the weight of the brain (mg) in 45 primate species. The line (regression) shows the expected brain weight for different body sizes. The X below the regression line indicates the expected brain size for humans; the actual size (represented by a red box) is much greater. Generally, prosimians are below the regression line, whereas anthropoids are above. This demonstrates that there was a selection pressure during human evolution for the development of brain tissue. Adapted from Stephan et al.7
In 1973, Jerison9 proposed that the proportion of brain to body size should take into account that as a simple matter of allometry, smaller organisms generally have larger brains, whereas larger ones tend to have proportionally smaller brains.
The development of any mammal brain is also subject to a series of minimum requirements. In principle, there must be a sufficient quantity of brain to maintain homeostasis (the internal equilibrium of the body in changing environmental conditions). There must also be a large enough quantity of brain and nervous tissue to enable perception of the outside world and the ability to respond appropriately to internal and external stimuli. These basic functions allow an animal to feed itself, flee from threats, and reproduce. Animals with more than this minimum amount of brain tissue necessary for survival have advantages over those that survive without exceeding these requirements. Therefore, small animals such as mice have relatively large brains in proportion to their body size, whereas animals such as elephants or whales, with larger bodies, have relatively small brains. A similar phenomenon occurs in organisms’ ontological development: compared to adult humans, babies have relatively large brains in proportion to body size. The same is true in primates: as an animal's body increases in size, the brain to body size ratio decreases. This is an example of allometric scaling. Both in absolute and in relative terms, it is evident that the brain of H. sapiens is larger than that of the chimpanzee, humans’ closest evolutionary relative.
It took several million years for the human brain to reach its current size. In comparison to primates as a whole, however, this process occurred very quickly. Some studies, such as those by Rakic and Kornack10 and Finlay and Darlington,11 suggest that the main mechanism giving rise to a larger brain occurs during embryonic development, with an increase in the production of the precursor cells that eventually form the brain (the neural tube).
Although the reasons for the accelerated evolution of the human brain remain unclear, it is apparent that this acceleration encompasses at least the entire Homo genus, continuing until the emergence of Neanderthal man and H. sapiens. According to Holloway et al.,12 the brain began to increase significantly in size in the australopiths (possibly the immediate predecessors to the genus Homo), whose body was similar in size to that of chimpanzees and whose brain increased in size by between 450 and probably 515cm3. The first members of the genus Homo (H. habilis, H. rudolfensis) had a brain volume of almost 700cm3, meaning the change that took place between them and H. ergaster and H. erectus, with a brain volume of approximately 1000cm3, was less dramatic.5
Structural reorganisation as part of the evolution of the brainIt is clear that increasing brain size, whether viewed in absolute or in relative terms, is not the only evolutionary change leading to the human brain. Holloway et al.12 argue that a restructuring of the tissues and circuits of the brain was essential in the evolution of the human brain. Larger brains require higher numbers of neurons and neuronal connections, which could eventually reduce brain efficiency; reorganisation is necessary to prevent this.13
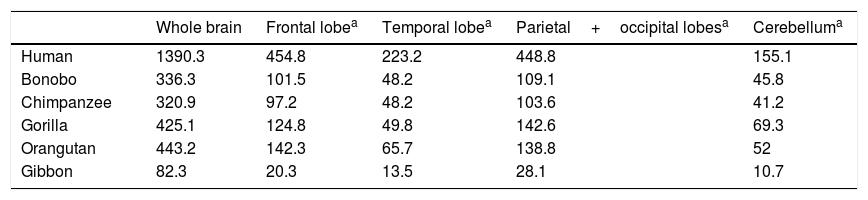
Semendeferi and Damasio14 used MRI to study the frontal lobe volumes of various apes and the modern human, reporting surprising results. Firstly, they found no great increase in size of the frontal lobe, which had been thought to be the main cause of encephalisation in humans. The human frontal lobe is larger in allometric terms, but not by much more than the margin that would be expected for a brain such as ours. However, while the total size of the human frontal lobe is no greater than that of a large-brained ape, certain parts do appear to have grown significantly in humans (Table 1).
Volume (cm3) of the brain and its different lobes in various primate species, according to results from 3-dimensional magnetic resonance imaging.
| Whole brain | Frontal lobea | Temporal lobea | Parietal+occipital lobesa | Cerebelluma | |
|---|---|---|---|---|---|
| Human | 1390.3 | 454.8 | 223.2 | 448.8 | 155.1 |
| Bonobo | 336.3 | 101.5 | 48.2 | 109.1 | 45.8 |
| Chimpanzee | 320.9 | 97.2 | 48.2 | 103.6 | 41.2 |
| Gorilla | 425.1 | 124.8 | 49.8 | 142.6 | 69.3 |
| Orangutan | 443.2 | 142.3 | 65.7 | 138.8 | 52 |
| Gibbon | 82.3 | 20.3 | 13.5 | 28.1 | 10.7 |
Taken from Semendeferi and Damasio.14
These researchers also discovered that the temporal lobes, known to be important in hearing, sight, and many other such higher cognitive functions as memory and language, were abnormally large in humans.14 This seems to suggest the existence of a selective pressure on the genus Homo, driving the development of this part of the brain.
Another new discovery from this important work was the relatively small size of the human cerebellum. This information is not fully consistent with Weaver's15 findings in a study including H. habilis, H. erectus, Neanderthal man, and early and modern H. sapiens. According to these results, there is a tendency for the cerebellum to decrease in relative size, but this pattern suddenly changes in modern H. sapiens, with the cerebellum growing once more. This would explain the remarkable cognitive capacity of modern humans compared not only to other members of the genus Homo, but also to early members of our own species.15
In an endocranial comparison of different Homo species, Bruner et al.16 found that the main difference between the human brain and that of the other species was the expansion of the parietal lobe, particularly the superior sections. The parietal lobe is one of the brain regions involved in processing syntax and grammar, highly important aspects of human language, and is the area that has undergone the greatest growth in H. sapiens.
In summary, while human encephalisation is strongly linked to increases in total and relative brain size, structural changes in specific brain regions and segments are also of undeniable importance in this process. While human frontal lobe growth was not as great as had previously been thought, other areas, particularly the temporal and parietal lobes, have undergone disproportionate growth.
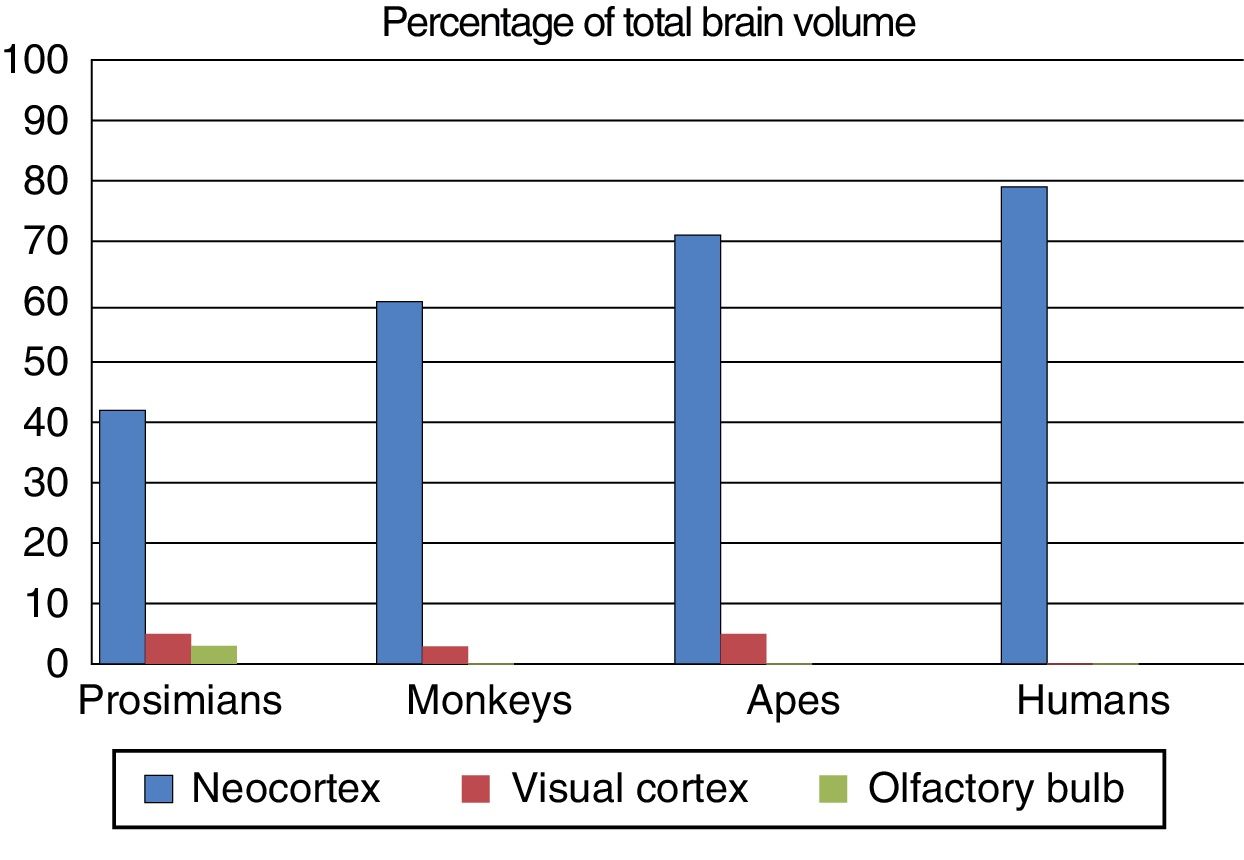
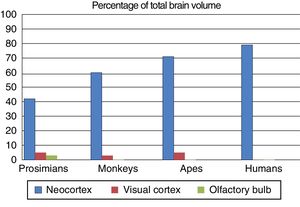
Fundamental adaptive change in the evolution of the brainThree main adaptive changes in brain morphology are known to have occurred during primate evolution: a reduction in the relative importance of olfaction; an increase in the relative importance of sight; and a sharp increase in the relative importance of the neocortex.17
These morphological changes in the brain were probably triggered by changes in the prevailing environmental conditions in the different evolutionary periods. Prosimians are generally small, and coveted by such predators as big cats and birds; it is for this reason that their primitive adaptation has maintained their nocturnal activity. Their communication, particularly when they are seeking sexual mates, uses olfactory signals such as urine and other secretions. This use and reliance on olfaction is evident in the large olfactory bulbs in these species.
On the other hand, no anthropoid primate species is nocturnal, with the exception of the South American night monkeys. Primates’ visual acuity and colour vision allows them to easily detect the colouration of a ripe fruit, or the red swelling of a potential mate; they therefore rely on visual information for success in their environment. Three-dimensional vision is a feature of great adaptive capacity, as primates lived in forests for a large part of their evolutionary history. The relative increase in the size and complexity of the visual cortex in primates, and particularly in anthropoids, owes to this adaptive change.
The third adaptive change is the absolute and relative increase in the size of the neocortex, which is involved in processing sensory information. During the evolution of the anthropoid brain, primates with more association areas for information processing were more likely to reproduce, transmitting their genes to their offspring. They were more intelligent, with more flexible behaviour and a greater capacity to adapt to environmental change. Fig. 3 shows the differing percentages of total brain size made up by these regions in different primate species.
The olfactory bulb, visual cortex, and neocortex as a percentage of total brain volume in prosimians, monkeys, apes, and modern humans. Adapted from Stephan et al.7
In the context of human evolution in general, it is important to understand the magnitude of the species’ leap in achieving “humanisation”. This term applies to a series of major and minor differences between H. sapiens and our closest evolutionary cousins, bonobos (Pan paniscus) and chimpanzees (P. troglodytes), from which humans diverged over 7 million years ago.18 These differences are evident from a morphological, cognitive, and cultural perspective; some of the most important differences determining the evolution of the brain have been analysed in the literature. Nonetheless, it should be noted that while genetically, humans and chimpanzees differ in only 1.23% of their genomes, this translates into an enormous phenotypic difference.19,20
Living creatures evolve, and this evolution enables them to adapt to a constantly changing environment. This requires 2 adaptation mechanisms: species-level genetic change, and individual-level changes mainly affecting chromatin organisation. These mechanisms occur independently of one another, and differences may be observed in them, depending on the species concerned. The changes that take place during a species’ development also rely on adaptive mechanisms, which require constant epigenetic modifications.
As has already been mentioned, human evolution is characterised by a rapid increase in the size and complexity of the brain. Decades of research have led to significant advances in the identification of anatomical and physiological characteristics of the human brain; this has been achieved through the use of molecular tools such as comparative genomics, which led to countless hypotheses about the evolution of the brain.
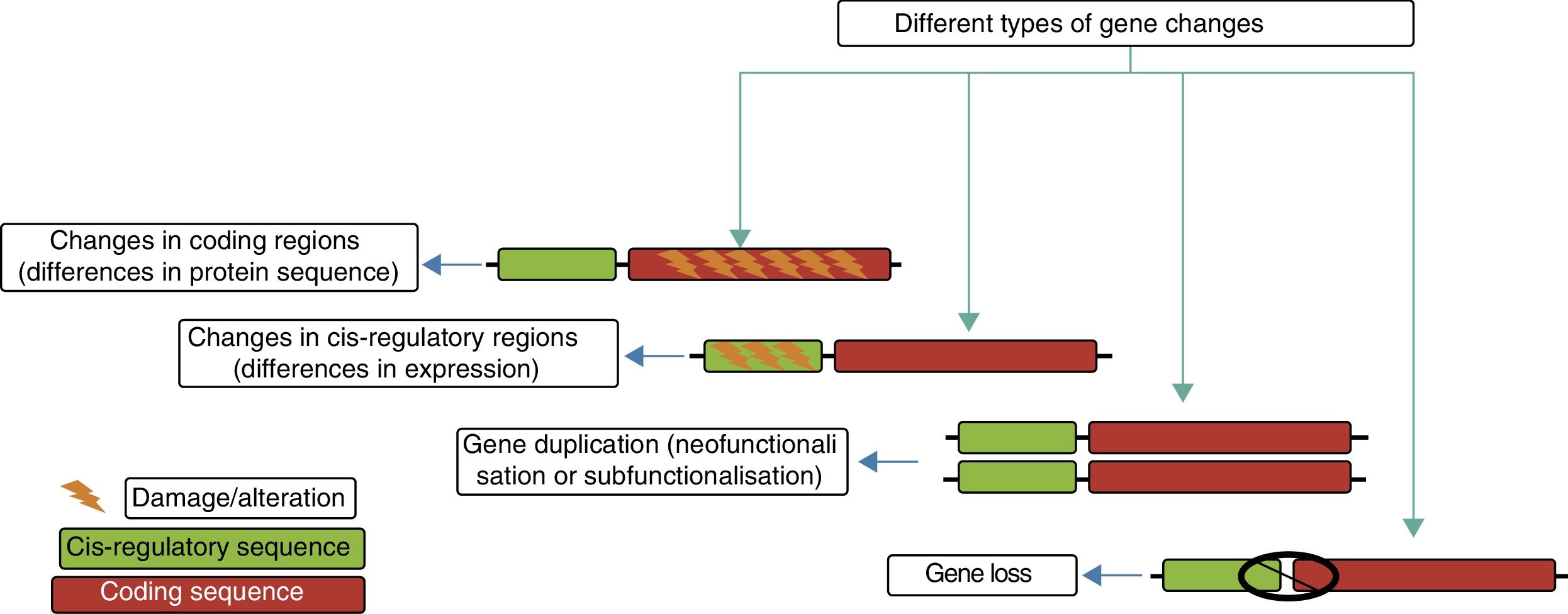
The genetic modifications that have led to the evolution of the human brain include epigenetic changes, single-nucleotide substitutions, and large-scale structural alterations in the genome. These genetic changes also have a great variety of functional consequences, from alterations in the amino acid sequence to changes in cis-regulatory regions, the creation of new genes, and the disappearance of existing ones.
Epigenetic modificationsAdaptive mechanisms largely involve so-called epigenetic mechanisms. Each adapting individual is “individualised” through interaction with the environment.10,20 It is thought that in humans, these epigenetic changes explain the additional 900g of grey matter and the distribution of the different areas of the brain.21 Without discussing the human genome in excessive detail, we should note that it contains between 20000 and 25000 genes, coding for all the proteins that constitute our biochemical structure. Protein coding sequences represent less than 2% of the genome; the other 98% is primarily made up of sequences regulating gene expression, including non-coding RNA sequences with regulatory functions (e.g. microRNA).20,21
As was previously mentioned, only 1.23% of the human genome differs from that of chimpanzees.22 In addition to the changes in protein structure and function, differences in gene expression patterns are thought to be fundamental in phenotypic diversification.23,24 Comparative studies have identified numerous transcriptional differences between humans and chimpanzees25; these are not exclusively genetic changes to regulatory elements (duplications, translocations, transversions, and transitions), but also include epigenetic modifications in regulatory regions.26
Although extensive research has been conducted into the genetic differences between humans and chimpanzees, these epigenetic differences have not been fully identified or characterised.27 The methylation of cytosines in CpG sites is known to be the main epigenetic modification in genomic DNA and can regulate the genetic expression of numerous coding sequences.28 In mammals, CpG sites containing methylated cytosines are characterised by high mutability. CpG islands are regions measuring 0.5-2kb and containing large numbers of CpG sites, and are generally located in non-methylated promoter regions of genes. The methylation of these regions is heritable through cell division; this is important in the maintenance of expression patterns.27 Variable levels of methylation are observed in regions adjacent to CpG islands during embryonic development, carcinogenesis, and cell reprogramming.29 Differences in methylation are also present in 12% to 18% of the genes that are expressed differently in humans and chimpanzees.27
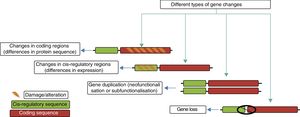
Genetic mechanisms in the evolution of the human brainThe principal genetic mechanisms that have been related to human brain evolution include: (1) positive selection in coding regions of the genome, leading to changes in the sequences of existing proteins; (2) the duplication and deletion of genes; and (3) changes caused by evolution in non-coding regions, particularly in cis-regulatory sequences, giving rise to modified genes (Fig. 4).
The different evolutionary changes observed in genes which may have contributed to human brain evolution. Adapted from Vallender et al.30
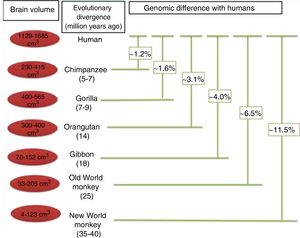
Protein sequence changes. Differences between protein sequences in humans and other mammals are numerous. A comparison between humans and chimpanzees shows that the majority of proteins in the 2 species have at least one amino acid difference. Some of these differences are probably of little importance functionally and in terms of the phenotypic evolution of the human species. However, other changes to amino acid sequences have undoubtedly had significant functional consequences, contributing to certain specific characteristics of the human brain. When positive selection acts on coding region mutations affecting a protein sequence, there is often an observable effect on the genes affected. The main focus of the study of the genetics, and particularly the evolution of the human brain, is the identification of genes that have shown positive selection compared to other primates (Fig. 5).30
Phylogenetic relationships between humans and other primates, indicating the temporal separation and degree of genetic difference between humans and the other species. The values shown for genetic difference represent nucleotide substitutions and do not account for other types of change (insertions, deletions, and structural changes). The values shown for Old and New World monkeys are based on groups of species within each clade. Adapted from Vallender et al.30
One field of research that has broadened our understanding of the evolution of the brain has been the identification of genes related to microcephaly, a congenital condition characterised by a considerable reduction in brain size. Among numerous manifestations, primary microcephaly presents as a reduction in brain size with no other abnormalities in brain architecture or gyral formation.30–32 The cerebral cortex is typically subject to a much greater reduction in size than other regions of the brain. Therefore, primary microcephaly could be considered an atavistic condition, recapitulating certain aspects of previous evolutionary stages of the human brain (smaller brain size, showing a disproportionate reduction in the cerebral cortex). Primary microcephaly is genetically heterogeneous. The condition has been related to 6 regions of the human genome (denominated microcephaly primary autosomal recessive 1 to 6 [MCPH1 to MCPH6]). Null mutations have also been found in 4 loci: microcephalin (MCPH1); CDK5 regulatory-subunit-associated protein 2 (CDK5RAP2); abnormal spindle-like, microcephaly-associated protein (ASPM); and centromeric protein J (CENPJ).30,33,34 The ASPM gene has undergone positive selection throughout the primate lineage to humans; this has been the case both in the lineage separating the great apes from the Old World monkeys and in the great ape lineage, including humans.30,35,36 The microcephalin gene is also characterised by a pronounced positive selection, primarily in the ancestral primate lineage leading to the great apes.35,37 The CDK5RAP2 and CENPJ genes display higher rates of non-synonymous substitutions in primates than in rodents; likewise, CDK5RAP2 shows particularly high mutation rates in human and chimpanzee lineages.38
An additional example related to the proliferation of neuronal precursor cells is adenylate-cyclase-activating polypeptide 1 (ADCYAP1), which has also undergone rapid protein-sequence evolution in humans. ADCYAP1 regulates the transition from proliferation to differentiated states in neurogenesis.39,40
Another area of neuronal development that may have been a substrate for positive selection in humans is axon guidance. The Abelson helper integration site 1 gene (AHI1) participates in directing axons from the brain to the spinal cord, and has been associated with Joubert syndrome, a rare brain disorder. The ASPM, ADCYAP1, and AHI1 genes all show high rates of non-synonymous sequence changes throughout human evolution, since the point at which humans diverged from chimpanzees.41 Another example is the Sonic Hedgehog (SHH) gene, which is highly conserved during development. The gene codes for a signal molecule which is key to the development of many types of tissue, particularly in the nervous and skeletal systems. The SHH protein comprises 2 functional domains: the signalling peptide and an autocatalytic region, which is responsible for cleaving the signalling peptide. The autocatalytic domain has been found to have a high rate of protein sequence evolution in primates, compared to other mammals. This effect is even more striking in humans. Interestingly, the main protein sequence changes observed involve gains of certain amino acids, such as serines and threonines, which are potential substrates of post-translational modifications. In view of this, SHH has been proposed as the main element in primate-to-human evolution of the nervous and/or skeletal systems.42 Finally, the gene monoamine oxidase A has also been related to these processes. This gene encodes a mitochondrial enzyme which catabolises various neurotransmitters, including dopamine, serotonin, and norepinephrine. Functional alterations in this gene may have numerous physiological and behavioural effects. It has been suggested that non-synonymous mutations in this gene may have caused functional changes to the enzyme, contributing to divergence between humans and chimpanzees.43
The case of the FOXP2 gene. Positive selection may also have occurred in genes related to language. The most significant example is the forkhead box P2 (FOXP2) gene; loss of function of this gene leads to a hereditary language disorder characterised by verbal dyspraxia. Interestingly, an association has been found between FOXP2 and verbal communication in other species, including mice and birds.30,44,45 Despite the very limited inter-species variation in the gene's coding sequence, human and chimpanzee FOXP2 genes are differentiated by 2 non-synonymous substitutions, which probably appeared less than 200000 years ago and are thought to have been involved in the origin of human language. The gene is also known to be involved in the development and function of various brain regions associated with language learning and production.45,46 Furthermore, and more significantly, FOXP2 controls fine motor tasks accompanying language production; point mutations in the gene are therefore thought to have contributed to linguistic fluency in humans.20
Creation of new genes. The creation of novel genes in the genome is triggered by duplication events, often within large gene families that may be predisposed to such events. The relaxation of evolutionary constraints on duplicate genes represents a unique opportunity for neofunctionalisation or subfunctionalisation.30 Neofunctionalisation involves the acquisition of new characteristics by one or both duplicate genes, while subfunctionalisation involves the ancestral function being partitioned between them.30
Recent advances made in the whole genome sequencing of several primate species, together with new technologies such as comparative genomic hybridisation, have enabled the identification of genes which have been duplicated over the course of human evolution.47–49 The first gene family in which substantial evidence has been found of gene duplication, followed by neofunctionalisation, is the Morpheus family. This gene family is so broad in human and great ape lineages that it was subject to intense positive selection on the encoded protein sequences. The function of the Morpheus genes remains to be determined; it is unclear whether the dramatic adaptation of the gene family through evolution is related to the process of human encephalisation.47
The MRG gene family, which codes for a group of G-protein–coupled receptors, is significantly expressed in the nociceptive neurons of the spinal cord; these genes are involved in modulating nociception.50 The presence of multiple copies of MRG genes in humans is probably due to the amplification of the gene after the human–mouse divergence: genetic sequencing of human copies of these genes reveals strong evidence of positive selection in the regions coding for extracellular ligand-binding domains.51,52
Another example of a duplication event with clear functional consequences is the creation of the glutamate dehydrogenase 2 (GLUD2) gene, which arose from the retrotransposition, or reintegration, of mRNA from a single ancestral precursor, GLUD1.53 In most mammal species, GLUD1 is the only gene coding for glutamate dehydrogenase, which catalyses recycling of the neurotransmitter glutamate in the brain.54,55 The retrotranscription event took place in the ape lineage after its diversion from the Old World monkeys; this gave rise to the GLUD2 gene, which codes for a different glutamate dehydrogenase protein, specific to apes and humans. Although GLUD1 is extensively expressed in many tissues, GLUD2 expression is exclusive to nervous and testicular tissue.54,55
Gene loss. Although the creation of new genes can have significant phenotypic consequences, gene loss in an organism can have dramatic, harmful effects, for which reason this is a much rarer evolutionary event. However, there are some cases where gene loss may occur due to selective changes over time. The best known example of gene loss in human evolution is in the olfactory receptor (OR) gene family.30,56,57 The mouse is thought to have approximately 1200 genes with OR functions, whereas humans have only around 350 of these genes.56 This reduction in the number of genes is due primarily to pseudogenes (non-functional relics of ancestral genes). The degeneration of the OR genes is not unique to humans, and appears also to affect several primate species. Positive selection has recently been demonstrated in several OR genes in the human lineage.57 Another interesting example of gene loss, which may have played a key role in the evolution of the human brain, is the myosin heavy chain 16 (MYH16) gene, encoding the myosin heavy chain 16 protein, which is present in skeletal muscle.58 In non-human primates, MYH16 is expressed only in muscles of the head, including those responsible for mastication. In humans, a mutation of this gene causes a frameshift mutation, resulting in loss of gene function. Humans have a relatively undeveloped mastication system compared to other primates; the loss of the MYH16 gene may have been partially responsible for this.59,60
Changes in gene expression. It has been proposed that changes in gene expression may have played an important part in the appearance of the human phenotype. In particular, some authors have argued that small changes in non-coding regulatory elements may have a strong impact on spatial and temporal expression patterns of developmental genes, with potentially profound phenotypic consequences.61–63
One strategy used to establish the role of changes in gene expression in human evolution is the comparison of the cis-regulatory regions of genes associated with the brain in order to identify which genes have been subject to positive selection. The prodynorphin (PDYN) gene codes for the precursor of an opioid neuropeptide involved in many neural processes. A cis-regulatory element located upstream of PDYN displays a high rate of sequence changes in the human lineage after human–chimpanzee divergence, demonstrating the effects of positive selection.64 In addition to cis-regulatory sequences, changes to the protein sequences of transcription factors can have a major influence over the expression of the genes that they regulate. Several studies have identified a significant increase in transcription factors among genes that have undergone positive selection in their protein coding regions; however, it is not clear whether this affects the expression patterns of their target genes.65,66
Non-coding RNAs. One interesting, fast-growing area of research is the identification of non-coding RNAs. In terms of methodology, this research is comparable to the studies that have aimed to identify evolutionary patterns in cis-regulatory elements. Consequently, many of the techniques developed to study the evolution of promoter or enhancer regions are applicable when studying the evolution of non-coding RNAs.67–70 In fact, during a large-scale scan for rapidly-evolving cis-regulatory elements, Pollard et al. found the first positively selected human non-coding RNA. This RNA is encoded by the human accelerated region 1 (HAR1) gene, which is expressed in the neurons of the developing human neocortex. The authors’ evolutionary analysis revealed that although the gene is only 118bp long, it contains 18 changes in the human lineage after human–chimpanzee divergence. Structural analysis of the HAR1 gene revealed that these changes affect the secondary structure of RNA in humans, compared with other amniotes, suggesting that the specific changes in the human HAR1 gene may have been involved in the evolution of the human cerebral cortex.67
Chromosomal differences between humans and the great apesThe genomic differences reported between humans and the great apes include chromosomal differences, changes in repeated DNA sequences, single-nucleotide polymorphisms, specific inactivation of genes, gene duplications and losses, etc.71 Chromosomal differences were the first observable genetic changes,72 making chromosome analysis an important tool in the study of genomic changes in evolutionary processes; the identification of ancestral chromosomal syntenies enables us to determine the evolutionary origin by phylogenetic line.73 It should be stressed that the X chromosome is considered the best preserved mammalian chromosome from an evolutionary perspective.74
With classic chromosome banding techniques, it was impossible to accurately establish homology or locate rearrangement breakpoints that occurred over the course of the evolution of humans and the great apes. However, this problem was largely solved in the 1980s with the introduction of new molecular probes such as fluorescence in situ hybridisation.73
Some chromosomal rearrangements between humans and chimpanzees may shed light on the speciation events that eventually led to the diversification of the 2 species.75 This approach enabled the analysis and characterisation of chromosomal material from such great apes as the chimpanzee, the gorilla, and the Sumatran orangutan subspecies (Pongo pygmaeus abelii), which led to the discovery that the human chromosome 2 is the result of the fusion of 2 homologous acrocentric chromosomes. The short arm (2p) originates in the chimpanzee and the long arm (2q) in the great apes; this fusion explains the modern human's chromosomal complement being reduced to 23 pairs.76–78 Furthermore, a pericentric inversion located on 2p reinforces the hypothesis of a phylogenetic link between humans and chimpanzees (independent ancestral condition), with this inversion taking place in a shared ancestor, subsequent to divergence from the gorilla. The gorilla and orangutan therefore conserve the ancestral forms.73,76,79
The main differences between the chromosomes of gorillas and chimpanzees and those of humans are a series of 11 rearrangements (9 para- and pericentric inversions, one translocation, and one fusion), differences in the nucleolus organiser regions, and additional G-bands in the subtelomeric regions. These subtelomeric modifications are composed of heterochromatin and are absent in humans and orangutans.80–82 While the majority of gorilla chromosomes show these caps, they are only present on half of chimpanzee chromosomes.83 These subtelomeric regions are known to be extremely dynamic areas.84 It has been suggested that genomic dynamism takes place when all chromatids interconnect (meiotic prophase), enabling exchange between the ends of non-homologous chromosomes.85 This may explain why subtelomeric regions associate preferentially and share a high degree of identical sequences located on non-homologous chromosomes.86 In this way, chromosome ends can be considered hotspots of genomic evolution.87
ConclusionsWe are yet to fully understand why the human brain is such a unique organ, why it is so special from the perspective of cognition, even compared with the brains of the higher primates and ancestral hominids, and when the brain truly became human. Nonetheless, the efforts of neurobiologists, biochemists, comparative anatomists, physiologists, and histologists, together with the analytical power of molecular genetics and genomics, have enabled us to better understand the complex, fascinating organ that is the human brain. Although we still do not fully understand the functioning of cognition, behaviour, or memory, the enormous efforts being made by countless researchers in all fields are gradually increasing our understanding of the most complex biological structure on the planet.
Conflicts of interestThe authors have no conflicts of interest to declare.
Please cite this article as: Rosales-Reynoso MA, Juárez-Vázquez CI, Barros-Núñez P. Evolución y genómica del cerebro humano. Neurología. 2018;33:254–265.