Alzheimer disease risk polymorphisms have been studied in patients with dementia, but have not yet been explored in mild cognitive impairment (MCI) in our population; nor have they been addressed in relation to cognitive variables, which can be predictive biomarkers of disease.
ObjectiveTo evaluate cognitive performance and presence of polymorphisms of the genes SORL1(rs11218304), PVRL2(rs6859), CR1(rs6656401), TOMM40(rs2075650), APOE (isoforms ε2, ε3, ε4), PICALM(rs3851179), GWAS_14q(rs11622883), BIN1(rs744373), and CLU(rs227959 and rs11136000) in patients with MCI and healthy individuals.
MethodologyWe performed a cross-sectional, exploratory, descriptive study of a prospective cohort of participants selected by non-probabilistic sampling, evaluated with neurological, neuropsychological, and genetic testing, and classified as cognitively healthy individuals and patients with MCI. Cognition was evaluated with the Neuronorma battery and analysed in relation to the polymorphic variants by means of measures of central tendency, confidence intervals, and nonparametric statistics.
ResultsWe found differences in performance in language and memory tasks between carriers and non-carriers of BIN1, CLU, and CR1 variants and a trend towards poor cognitive performance for PICALM, GWAS_14q, SORL1, and PVRL2 variants; the APOE and TOMM40 variants were not associated with poor cognitive performance.
DiscussionDifferences in cognitive performance associated with these polymorphic variants may suggest that the mechanisms regulating these genes could have an effect on cognition in the absence of dementia; however, this study was exploratory and hypotheses based on these results must be explored in larger samples.
Los polimorfismos de riesgo para el desarrollo de enfermedad de Alzheimer se han estudiado en pacientes con demencia, pero aún no se han explorado en trastorno neurocognitivo leve (TNL) en nuestra población, ni se han considerado en relación con variables cognitivas, las cuales pueden ser biomarcadores predictivos de enfermedad.
ObjetivoEvaluar los desempeños cognitivos y los polimorfismos en los genes SORL1(rs11218304), PVRL2(rs6859), CR1(rs6656401), TOMM40(rs2075650), APOE(isoformas ε2, ε3, ε4), PICALM(rs3851179), GWAS_14q(rs11622883), BIN(rs744373), CLU (rs227959 y rs11136000) en pacientes con TNL y en sujetos sanos.
MetodologíaEstudio descriptivo, exploratorio y transversal, en una cohorte prospectiva de participantes seleccionados mediante muestreo no probabilístico, evaluados por neurología, neuropsicología y genética, y clasificados como cognitivamente sanos y pacientes con TNL, según criterios. La cognición se evaluó por medio de la batería Neuronorma y se analizó en relación con las variantes polimórficas por medio de medidas de tendencia, intervalos de confianza y estadísticos no paramétricos.
ResultadosSe identificaron diferencias en los desempeños en tareas de lenguaje y memoria en relación con las variantes de BIN1, CLU y CR1, junto con tendencias en las variantes de PICALM, GWArs, SORL y PVRL2, mientras que en APOE y TOMM40 no se encontraron tendencias.
DiscusiónLas tendencias en los desempeños cognitivos en relación con variantes polimórficas podrían indicar que, en ausencia de demencia, los mecanismos que regulan estos genes podrían tener un efecto sobre la cognición; sin embargo, esta aproximación tiene un carácter exploratorio y sus resultados permiten generar hipótesis que requieren ser exploradas en muestras de mayor tamaño.
Mild cognitive impairment (MCI) may present with a wide range of symptoms, has multiple aetiologies, and may progress to major neurocognitive disorder; therefore, it has been suggested that it may constitute a transitional state between normal ageing and dementia.1 Few studies have analysed genetic risk factors for MCI. However, as patients with MCI present greater risk of developing such other neurocognitive disorders as Alzheimer disease (AD),2 it has been suggested that MCI may have a genetic component3; therefore, AD risk polymorphisms may be studied in these patients.
The genes associated with AD are linked to different pathophysiological mechanisms involving lipid metabolism, oxidative stress, the immune response, and transport and degradation mechanisms. The APOE gene is involved in lipid transport; this gene encodes apolipoprotein E, a plasma lipoprotein involved in maintaining the structure of the cell membrane and regulating cholesterol transport.4TOMM40 encodes one protein of a protein complex involved in protein import into mitochondria. The gene has been found to colocalise with amyloid precursor protein (APP) in mitochondria in the brains of patients with AD. Furthermore, carriers of the APOE ε3/4 genotype display APP accumulation, which supports the hypothesis that APOE is involved in APP transport into mitochondria and interacts with TOMM40 to facilitate import. TOMM40 has therefore been proposed as a candidate gene for AD.5
SORL1 is involved in transport and degradation. The gene encodes a receptor associated with protein transport, which directs phosphatases, kinases, and signalling receptors to their correct locations within the cell, ensuring proper cell and tissue function.6BIN1 also plays a role in protein transport and degradation. It encodes an adaptor protein involved in synaptic vesicle endocytosis and recovery; isoforms participate in apoptosis, inflammation, and calcium homeostasis.7PICALM, in turn, encodes an essential protein for intracellular trafficking of neurotransmitters, proteins, and lipids.3,8
CR1 has been associated with immune response mechanisms; the gene encodes a glycoprotein that facilitates antigen removal, triggers the inflammatory response, and mediates the clearance of amyloid β particles.9 Another gene involved in the immune response is PVRL2, which encodes a membrane glycoprotein that serves as an entrance for certain viral strains and is involved in cell-to-cell transmission of viruses.10 Lastly, CLU encodes a chaperone protein involved in protein folding and cell apoptosis.11
Some studies have analysed the association between some of these genes and cognitive function in patients with AD, MCI, and healthy individuals,12 as some gene variants are known to have an effect on cognitive function. Therefore, cognitive variables constitute a useful tool for detecting new risk loci and may serve as therapeutic targets and predictive biomarkers of the disease.13 For instance, some APOE variants have been associated with improved cognitive function,14 whereas the ε4 allele is associated with poorer episodic memory.15 Some TOMM40 variants are associated with improved cognitive performance, whereas others, such as poly-T length polymorphisms, are linked to poorer verbal learning and executive function.16 Furthermore, BIN1 genotype has been found to have an impact on working memory.17
In our setting, APOE explains only part of the risk of AD18; therefore, other single-nucleotide polymorphisms (SNP) associated with risk of the disease have been explored, and a significant association between risk of AD and a TOMM40 variant (rs2075650) has been reported. Some alleles of TOMM40 and the APOE 4 allele have been associated with onset of AD up to 13 years earlier.19 To date, no studies have evaluated AD risk variants in patients with MCI in our setting. Our purpose was to study the association between cognitive function and polymorphisms SORL1 rs11218304, PVRL2 rs6859, CR1 rs6656401, TOMM40 rs2075650, APOE rs7412 and rs440446, PICALM rs3851179, GWAS_14q rs11622883, BIN1 rs744373, and CLU rs227959 and rs11136000 in patients with MCI and cognitively healthy individuals.
MethodsWe conducted an exploratory, observational, descriptive, longitudinal study; therefore, our results cannot be extrapolated to the general population. The study was led by the neurosciences group and approved by the research ethics committee of the Faculty of Medicine at Universidad Nacional de Colombia. All participants signed informed consent forms.
Sample selectionParticipants were recruited by non-probability sampling between December 2015 and November 2017. We established the following inclusion criteria: age equal to or greater than 50 years; at least one year of schooling; not meeting diagnostic criteria for major neurocognitive disorder; no history of confusional syndrome, traumatic brain injury, cerebrovascular disease, or any other disorder that may cause cognitive alterations; and presenting no sensory limitations that may interfere with the participant's ability to complete the tests. The inclusion criteria were applied by a group of healthcare professionals specialising in neurocognitive assessment.
ProcedureClinical and sociodemographic variables were gathered during the neurological examination using a structured questionnaire. We used the following screening tools: Montreal Cognitive Assessment (MoCA),20 INECO Frontal Screening (IFS),21 Geriatric Depression Scale (GDS),22 and Neuropsychiatric Inventory (NPI).23 To assess functional independence we used a modified version of the Lawton Instrumental Activities of Daily Living (IADL) scale24 and the Spanish-language version of the Bayer Activities of Daily Living (B-ADL) scale.25 Cognitive function was evaluated with the NEURONORMA neuropsychological test battery, normalised to our population.26,27 The test battery evaluates 5 cognitive domains: 1) Attention: parts A and B of the Trail Making Test (TMT), Symbol Digit Modalities Test (SDMT), Stroop Color-Word Test, verbal and visual span forward; 2) Memory: Free and Cued Selective Reminding Test (FCSRT), Rey-Osterrieth Complex Figure Test (recall); 3) Language: Boston Naming Test (BNT), Token Test, verbal fluency test; 4) Constructional ability: Rey-Osterrieth Complex Figure Test (copy); and 5) Executive function: phonemic verbal fluency, Tower of London test, Stroop Color-Word Test, Wisconsin Card Sorting Test.
Participants were considered to be cognitively healthy if they performed as expected on the screening tests, with scores being a maximum of 1 SD below the mean for their age and education level.26,27 MCI was diagnosed according to the criteria established in the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5).28 We studied the following SNPs: SORL1 rs11218304, PVRL2 rs2075650, CR1 rs6656401, TOMM40 rs2075650, APOE rs7412 and rs440446, PICALM rs3851179, GWAS_14q rs11622883, BIN rs744373, and CLU rs227959 and rs11136000. DNA was extracted from peripheral blood samples using the ReliaPrep Blood gDNA Miniprep System (Promega, USA). Genotyping was performed using TaqMan probes (Applied Biosystems, USA); this technique enables discrimination between 2 alleles of a specific SNP. Each assay contains 2 primers (forward and reverse), a VIC® dye–labelled MGB probe (which detects the sequence of an allele), and a 6-FAM™ dye–labelled MGB probe (which detects the other allele). Reactions were run with the Bio-Rad CFX96 Touch system, under the following conditions: 50°C for 2minutes, 95°C for 10minutes, 50 cycles at 95°C for 15seconds, and 60°C for 1minute. Allelic discrimination was performed using Bio-Rad CFX Manager software.
Data analysisClinical and sociodemographic characteristics are expressed with absolute and relative frequencies or measures of central tendency and dispersion, depending on the type of data. The results of the psychometric assessment were converted to scaled scores according to the corresponding percentile in the general population.
Psychometric test results were analysed by patient subgroups established according to polymorphic variants; results are expressed as relative frequencies. We also obtained 95% confidence intervals (CI) using percentiles 2.5 and 97.5 of the bootstrap distribution, selecting 1000 random samples with 75% replacement.29
Demographic, cognitive, and genetic variables were analysed with the non-parametric Mann-Whitney U test.29 The association between allele frequencies and diagnosis was evaluated with the Fisher exact test. We used the Kruskal-Wallis test to detect any differences in cognitive function between patients with different polymorphic variants. We also analysed the frequencies of good, average, borderline, and poor cognitive function for each allele pair by visually analysing a table of absolute and relative frequencies. Frequencies showing a trend towards statistical significance for some specific tests and consistency below test performance were evaluated by comparing 95% CIs to determine whether the differences were relevant and may be associated with any of the results of non-parametric tests.
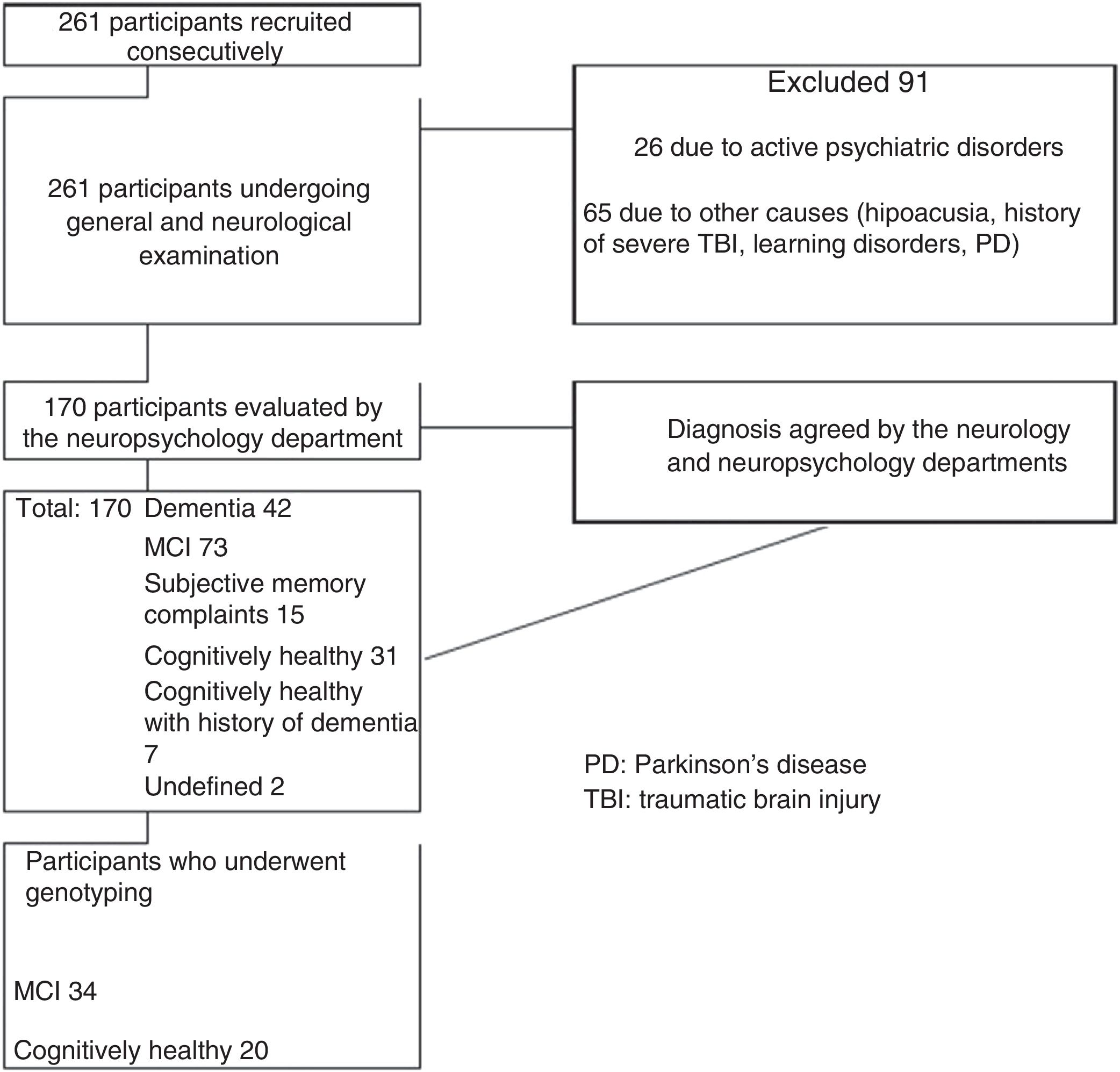
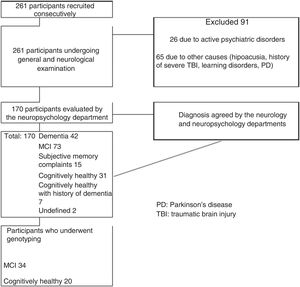
ResultsOf the initial sample of 261 participants, 91 were excluded for not meeting the inclusion criteria. Thirty-one participants (11.8%) were cognitively healthy individuals with no family history of dementia, 20 of whom underwent genotyping, and 73 individuals (28%) had MCI, 34 of whom underwent genotyping. Our final sample included 20 cognitively healthy controls and 34 patients with MCI. Fig. 1 summarises the patient selection process.
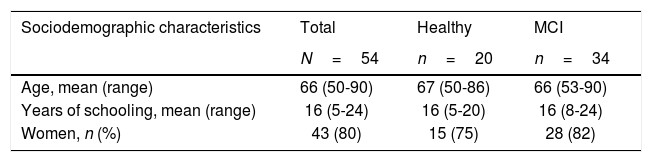
Both groups showed a greater proportion of women (MCI: 82.35%; 95% CI, 69.5%-95%; controls: 75%; 95% CI, 56%-94%). Mean age was 66 years (range, 53-90) among patients with MCI and 67 years (range, 50-86) among controls. In both groups, participants had a minimum of 5 years of schooling. Dyslipidaemia was the most frequent cardiovascular risk factor in both groups, and diabetes mellitus was the least frequent. Table 1 summarises the clinical and sociodemographic characteristics of our sample.
Sociodemographic and clinical characteristics of the sample.
| Sociodemographic characteristics | Total | Healthy | MCI |
|---|---|---|---|
| N=54 | n=20 | n=34 | |
| Age, mean (range) | 66 (50-90) | 67 (50-86) | 66 (53-90) |
| Years of schooling, mean (range) | 16 (5-24) | 16 (5-20) | 16 (8-24) |
| Women, n (%) | 43 (80) | 15 (75) | 28 (82) |
| Clinical characteristics | Total | Controls | MCI | |||
|---|---|---|---|---|---|---|
| N=54 | 95% CI | n=20 | 95% CI | n=34 | 95% CI | |
| Arterial hypertension, n (%) | 17 (31.5) | 21.1-49.9 | 6 (30) | 13.1-46.9 | 11 (32) | 19.1-45.6 |
| Diabetes mellitus, n (%) | 8 (15) | 6.8-22.8 | 2 (10) | 0-21.1 | 6 (17) | 6.9-29.4 |
| Dyslipidaemia, n (%) | 28 (52) | 40.6-63.1 | 10 (50) | 31.6-68.4 | 18 (53) | 38.8-67.1 |
| Smoking, n (%) | 15 (27) | 17.7-37.8 | 4 (20) | 5.2-34.8 | 11 (32) | 19.1-45.6 |
| Sedentary lifestyle, n (%) | 11 (20) | 11.3-29.4 | 6 (30) | 13.1-46.9 | 5 (15) | 4.7-24.7 |
CI: confidence interval.
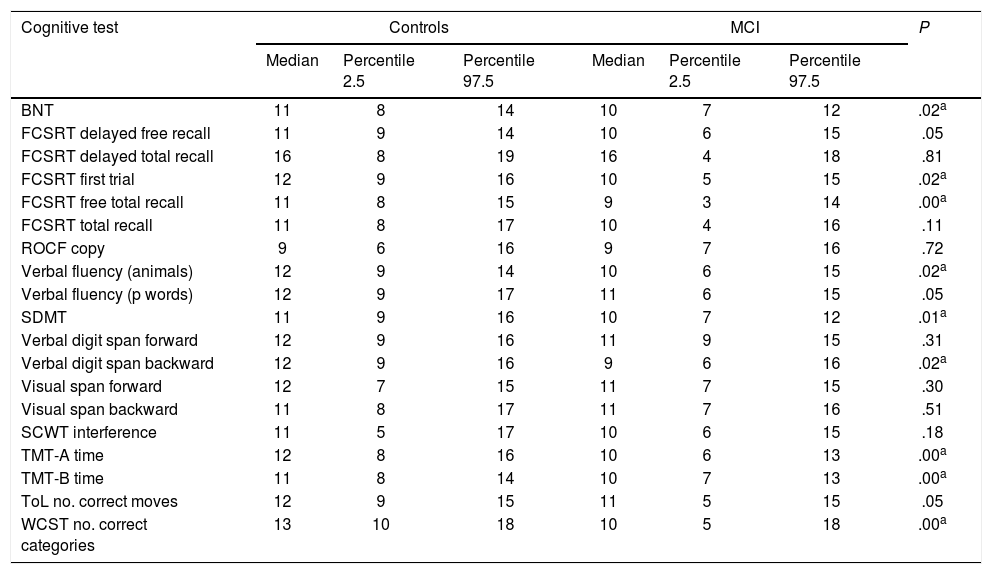
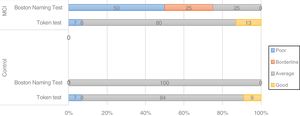
Screening tests identified significant differences between groups. Controls scored a median of 25 points (95% CI, 20-28) on the MoCA and 27 (95% CI, 24-30) on the IFS, whereas patients with MCI scored 23 (95% CI, 17-27) and 26 points (95% CI, 21-29), respectively (P=.02). In the NEURONORMA test battery, we observed high scaled scores (Table 2) and significant differences between groups in most tasks, except for the Rey-Osterrieth Complex Figure Test (copy), the Stroop Color-Word Test (interference), short-term memory tasks (verbal and visual span forward), working memory tasks (visual span backward), and cued recall tasks.
Cognitive performance on the NEURONORMA test battery (scaled scores).
| Cognitive test | Controls | MCI | P | ||||
|---|---|---|---|---|---|---|---|
| Median | Percentile 2.5 | Percentile 97.5 | Median | Percentile 2.5 | Percentile 97.5 | ||
| BNT | 11 | 8 | 14 | 10 | 7 | 12 | .02a |
| FCSRT delayed free recall | 11 | 9 | 14 | 10 | 6 | 15 | .05 |
| FCSRT delayed total recall | 16 | 8 | 19 | 16 | 4 | 18 | .81 |
| FCSRT first trial | 12 | 9 | 16 | 10 | 5 | 15 | .02a |
| FCSRT free total recall | 11 | 8 | 15 | 9 | 3 | 14 | .00a |
| FCSRT total recall | 11 | 8 | 17 | 10 | 4 | 16 | .11 |
| ROCF copy | 9 | 6 | 16 | 9 | 7 | 16 | .72 |
| Verbal fluency (animals) | 12 | 9 | 14 | 10 | 6 | 15 | .02a |
| Verbal fluency (p words) | 12 | 9 | 17 | 11 | 6 | 15 | .05 |
| SDMT | 11 | 9 | 16 | 10 | 7 | 12 | .01a |
| Verbal digit span forward | 12 | 9 | 16 | 11 | 9 | 15 | .31 |
| Verbal digit span backward | 12 | 9 | 16 | 9 | 6 | 16 | .02a |
| Visual span forward | 12 | 7 | 15 | 11 | 7 | 15 | .30 |
| Visual span backward | 11 | 8 | 17 | 11 | 7 | 16 | .51 |
| SCWT interference | 11 | 5 | 17 | 10 | 6 | 15 | .18 |
| TMT-A time | 12 | 8 | 16 | 10 | 6 | 13 | .00a |
| TMT-B time | 11 | 8 | 14 | 10 | 7 | 13 | .00a |
| ToL no. correct moves | 12 | 9 | 15 | 11 | 5 | 15 | .05 |
| WCST no. correct categories | 13 | 10 | 18 | 10 | 5 | 18 | .00a |
Values represent scaled scores on each cognitive test normalised to our population.
BNT: Boston Naming Test; FCSRT; Free and Cued Selective Reminding Test; MCI: mild cognitive impairment; ROCF: Rey-Osterrieth Complex Figure test; SCWT: Stroop Color-Word Test; SDMT: Symbol Digit Modalities Test; TMT: Trail Making Test; ToL: Tower of London; WCST: Wisconsin Card Sorting Test.
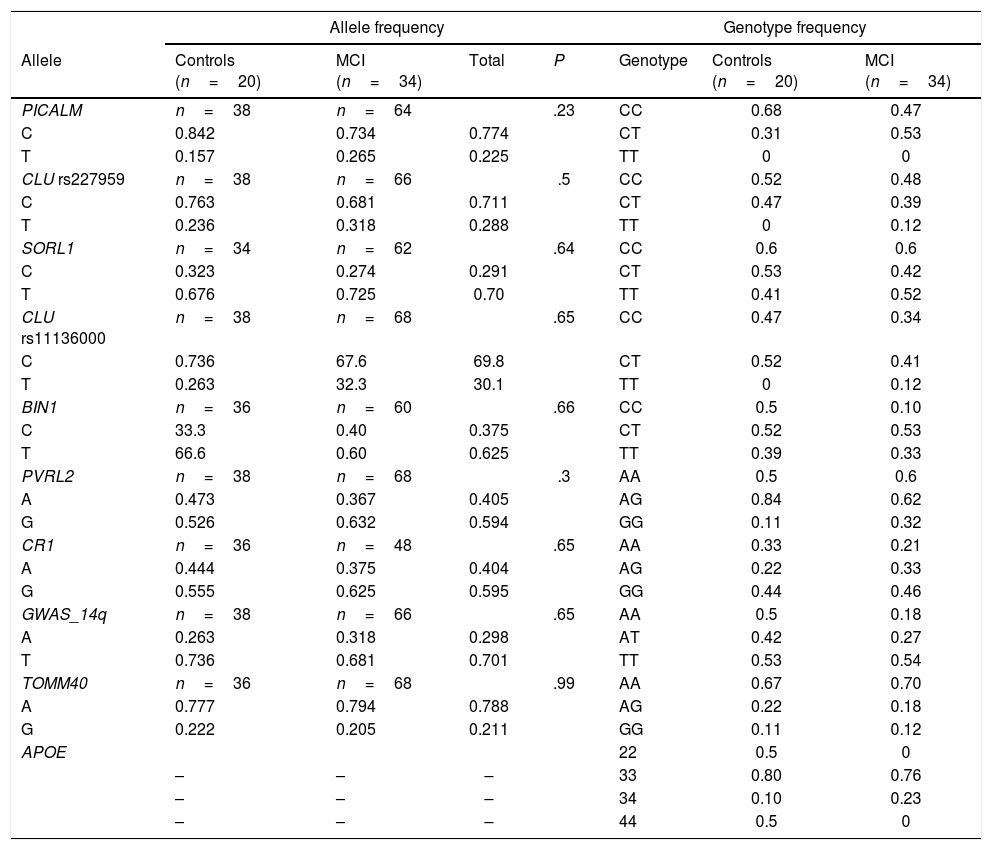
All polymorphisms were in Hardy-Weinberg equilibrium, with the exception of PVRL2 rs6859 in the MCI group. No significant associations were observed between polymorphisms and diagnosis. Allele and genotype frequencies are shown in Table 3. We compared cognitive performance according to presence of different polymorphisms using non-parametric statistics.
Distribution of allele and genotype frequencies.
| Allele frequency | Genotype frequency | ||||||
|---|---|---|---|---|---|---|---|
| Allele | Controls (n=20) | MCI (n=34) | Total | P | Genotype | Controls (n=20) | MCI (n=34) |
| PICALM | n=38 | n=64 | .23 | CC | 0.68 | 0.47 | |
| C | 0.842 | 0.734 | 0.774 | CT | 0.31 | 0.53 | |
| T | 0.157 | 0.265 | 0.225 | TT | 0 | 0 | |
| CLU rs227959 | n=38 | n=66 | .5 | CC | 0.52 | 0.48 | |
| C | 0.763 | 0.681 | 0.711 | CT | 0.47 | 0.39 | |
| T | 0.236 | 0.318 | 0.288 | TT | 0 | 0.12 | |
| SORL1 | n=34 | n=62 | .64 | CC | 0.6 | 0.6 | |
| C | 0.323 | 0.274 | 0.291 | CT | 0.53 | 0.42 | |
| T | 0.676 | 0.725 | 0.70 | TT | 0.41 | 0.52 | |
| CLU rs11136000 | n=38 | n=68 | .65 | CC | 0.47 | 0.34 | |
| C | 0.736 | 67.6 | 69.8 | CT | 0.52 | 0.41 | |
| T | 0.263 | 32.3 | 30.1 | TT | 0 | 0.12 | |
| BIN1 | n=36 | n=60 | .66 | CC | 0.5 | 0.10 | |
| C | 33.3 | 0.40 | 0.375 | CT | 0.52 | 0.53 | |
| T | 66.6 | 0.60 | 0.625 | TT | 0.39 | 0.33 | |
| PVRL2 | n=38 | n=68 | .3 | AA | 0.5 | 0.6 | |
| A | 0.473 | 0.367 | 0.405 | AG | 0.84 | 0.62 | |
| G | 0.526 | 0.632 | 0.594 | GG | 0.11 | 0.32 | |
| CR1 | n=36 | n=48 | .65 | AA | 0.33 | 0.21 | |
| A | 0.444 | 0.375 | 0.404 | AG | 0.22 | 0.33 | |
| G | 0.555 | 0.625 | 0.595 | GG | 0.44 | 0.46 | |
| GWAS_14q | n=38 | n=66 | .65 | AA | 0.5 | 0.18 | |
| A | 0.263 | 0.318 | 0.298 | AT | 0.42 | 0.27 | |
| T | 0.736 | 0.681 | 0.701 | TT | 0.53 | 0.54 | |
| TOMM40 | n=36 | n=68 | .99 | AA | 0.67 | 0.70 | |
| A | 0.777 | 0.794 | 0.788 | AG | 0.22 | 0.18 | |
| G | 0.222 | 0.205 | 0.211 | GG | 0.11 | 0.12 | |
| APOE | 22 | 0.5 | 0 | ||||
| – | – | – | 33 | 0.80 | 0.76 | ||
| – | – | – | 34 | 0.10 | 0.23 | ||
| – | – | – | 44 | 0.5 | 0 | ||
MCI: mild cognitive impairment. Values are expressed as percentages.
P values below .05 are considered statistically significant (Fisher exact test).
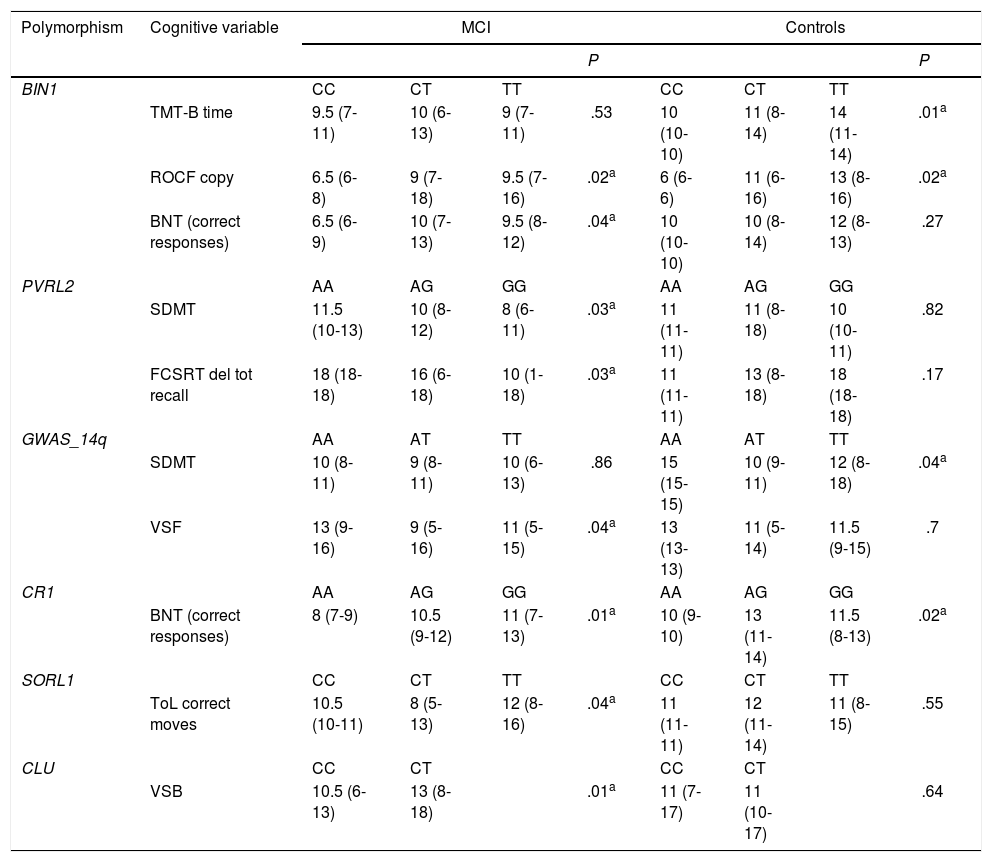
In patients with MCI, non-parametric tests (Table 4) showed significant differences in language (BNT) associated with variants of BIN1 (P=.04) and CR1 (P=.01), differences in attention (SDMT) and memory (FCSRT–delayed total recall) associated with PVRL2 variants (P=.03), and differences in visual span forward score associated with GWAS_14q variants (P=.04). We also observed significant differences in executive function associated with SORL and CLU variants (Tower of London [P=.04] and visual span backward [P=.01], respectively). Controls presented differences in attention associated with variants of GWAS_14q and BIN1 (SDMT [P=.04] and TMT-B [P=.01], respectively), and differences in language associated with CR1 variants (BNT [P=.01]).
Cognitive tests displaying significant differences associated with genotypic variants.
| Polymorphism | Cognitive variable | MCI | Controls | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | P | ||||||||
| BIN1 | CC | CT | TT | CC | CT | TT | |||
| TMT-B time | 9.5 (7-11) | 10 (6-13) | 9 (7-11) | .53 | 10 (10-10) | 11 (8-14) | 14 (11-14) | .01a | |
| ROCF copy | 6.5 (6-8) | 9 (7-18) | 9.5 (7-16) | .02a | 6 (6-6) | 11 (6-16) | 13 (8-16) | .02a | |
| BNT (correct responses) | 6.5 (6-9) | 10 (7-13) | 9.5 (8-12) | .04a | 10 (10-10) | 10 (8-14) | 12 (8-13) | .27 | |
| PVRL2 | AA | AG | GG | AA | AG | GG | |||
| SDMT | 11.5 (10-13) | 10 (8-12) | 8 (6-11) | .03a | 11 (11-11) | 11 (8-18) | 10 (10-11) | .82 | |
| FCSRT del tot recall | 18 (18-18) | 16 (6-18) | 10 (1-18) | .03a | 11 (11-11) | 13 (8-18) | 18 (18-18) | .17 | |
| GWAS_14q | AA | AT | TT | AA | AT | TT | |||
| SDMT | 10 (8-11) | 9 (8-11) | 10 (6-13) | .86 | 15 (15-15) | 10 (9-11) | 12 (8-18) | .04a | |
| VSF | 13 (9-16) | 9 (5-16) | 11 (5-15) | .04a | 13 (13-13) | 11 (5-14) | 11.5 (9-15) | .7 | |
| CR1 | AA | AG | GG | AA | AG | GG | |||
| BNT (correct responses) | 8 (7-9) | 10.5 (9-12) | 11 (7-13) | .01a | 10 (9-10) | 13 (11-14) | 11.5 (8-13) | .02a | |
| SORL1 | CC | CT | TT | CC | CT | TT | |||
| ToL correct moves | 10.5 (10-11) | 8 (5-13) | 12 (8-16) | .04a | 11 (11-11) | 12 (11-14) | 11 (8-15) | .55 | |
| CLU | CC | CT | CC | CT | |||||
| VSB | 10.5 (6-13) | 13 (8-18) | .01a | 11 (7-17) | 11 (10-17) | .64 | |||
Data are expressed as median (range). BNT: Boston Naming Test; FCSRT del tot recall: Free and Cued Selective Reminding Test–delayed total recall; MCI: mild cognitive impairment; ROCF: Rey-Osterrieth Complex Figure test; SDMT: Symbol Digit Modalities Test; TMT: Trail Making Test; VSB: visual span backward; VSF: visual span forward.
Values represent scaled scores on each cognitive test normalised to our population.
We used scaled scores to establish 4 levels of cognitive performance (poor: < 6; borderline: 7; average: 8-13; good: > 14) and analysed the frequency of each level in subgroups established according to genetic variants. We present the results that showed consistency between non-parametric tests and frequency analysis.
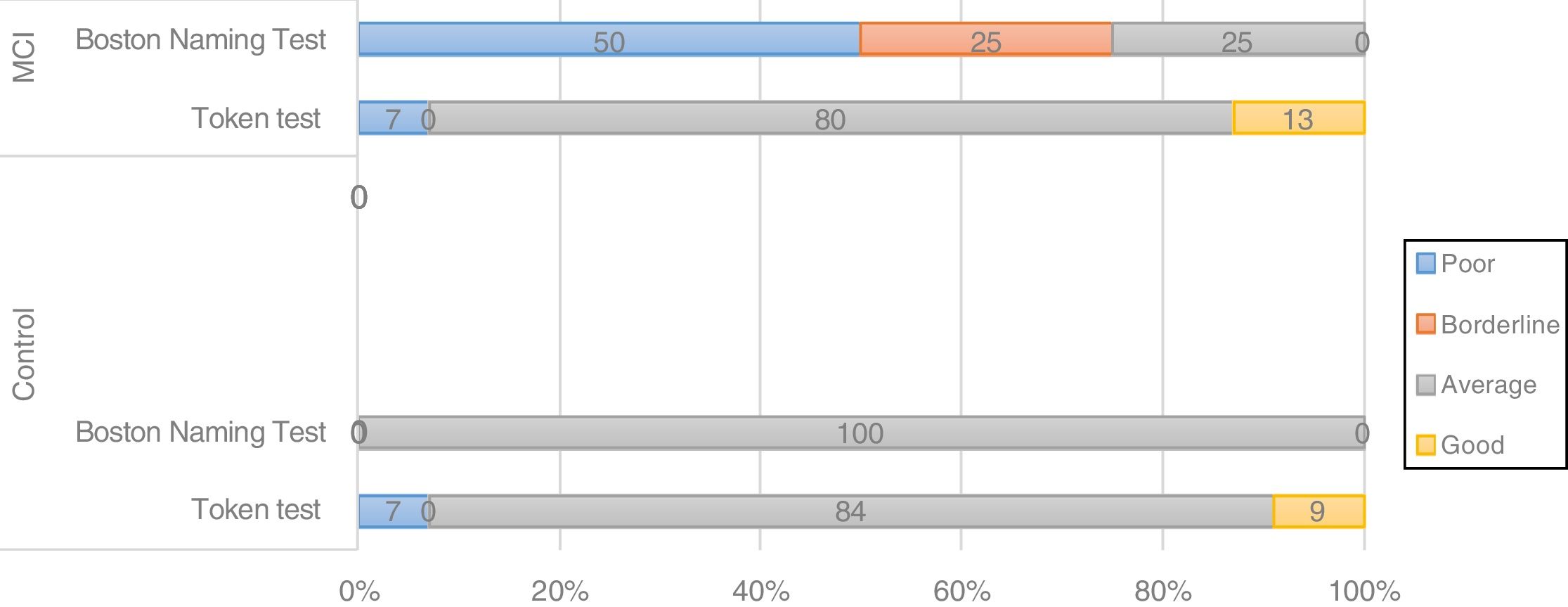
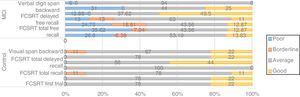
The BIN1 CC genotype was associated with differences in the distribution of cognitive performance groups between controls and patients with MCI; 50% of patients with the CC genotype showed poor performance and 25% showed borderline performance in a language task (BNT) (Fig. 2), whereas carriers of genotypes CT and TT showed good performance in 80% and 100% of cases, respectively. In controls, all BIN1 variants were associated with good or average cognitive performance in 100% of cases, which suggests that the CC genotype may be a risk variant in patients with MCI.
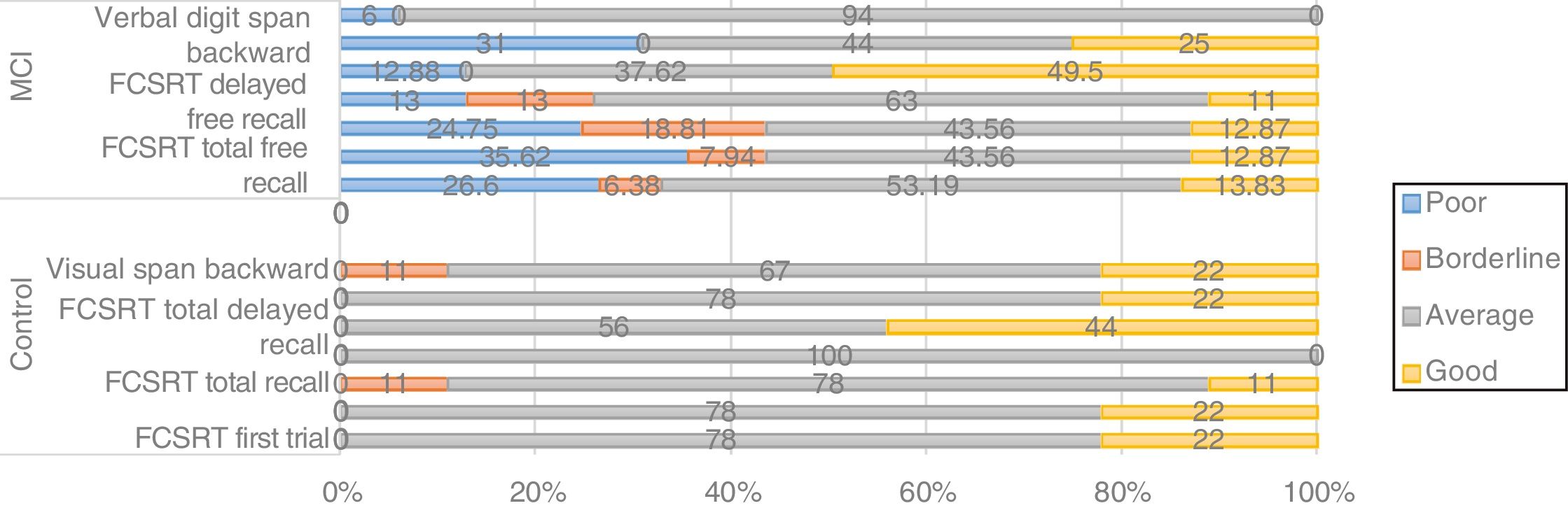
Thirty-one percent of patients carrying the CC genotype of CLU (rs11136000) showed poor or borderline performance in memory tasks (Fig. 3). Furthermore, non-parametric comparisons also detected differences in performance in the visual span backward. We may therefore hypothesise that certain genotypes of CLU (rs11136000) may be associated with memory performance.
Regarding CR1 in patients with MCI, nearly half of carriers of the GG variant performed poorly in 2 memory tasks (46% performed poorly in FCSRT–total recall and FCSRT–free total recall). Non-parametric tests detected differences in BNT performance associated with CR1 variants, both in patients with MCI and in controls; this is consistent with the results of the frequency analysis, since the BNT involves semantic memory.
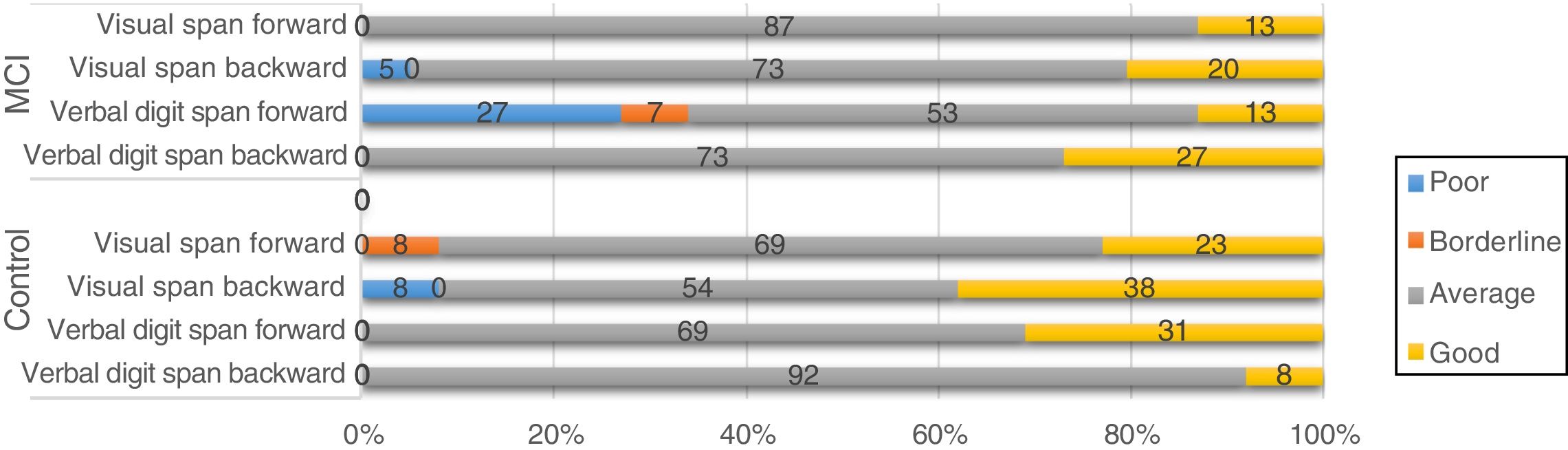
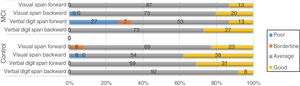
Regarding the genes SORL, PVRL2, GWAS_14q, and PICALM, although frequency analysis showed an association between variants and performance in some tasks (eg, PICALM genotype CC was associated with poor performance in a working memory task; Fig. 4), these results were not consistent with those of the non-parametric tests. Furthermore, confidence intervals were wide, which suggests that the differences found may be due to the variability of cognitive function in this type of patients. No association was observed between APOE and TOMM40 polymorphisms and cognitive performance (results not shown).
DiscussionWe explored different gene variants and cognitive profiles in a group of patients with MCI, given that these individuals present considerable risk of developing major neurocognitive disorders, such as AD, and that analysing cognitive performance associated with specific gene variants may assist in prevention research.13 Our study found an association between cognitive performance and some polymorphisms; while these results are merely exploratory, due to the small size of our sample, they suggest an interaction between cognitive and genetic variables that should be analysed in future studies with larger samples.
In the case of BIN1, an association was observed between the CC genotype and cognitive performance in a language task (BNT); the association was also observed in non-parametric data. BIN1 is the second most important risk locus for late-onset Alzheimer disease, after APOE,30 and has been associated with poorer cognitive performance in young healthy individuals,17,30 which supports the hypothesis that it may affect such clinical variables as cognitive function.
The CR1 rs6656401 GG genotype is reported to be associated with global cognitive impairment, episodic memory impairment both in patients with late-onset Alzheimer disease and in cognitively healthy individuals,31 amyloid deposition,32 and poor semantic memory, visuospatial ability, and perceptual speed.9 Our study found an association between this polymorphism and performance on the BNT; our results suggest that this tool may be useful for evaluating patients with MCI and analysing the association between MCI and certain biomarkers.
Few studies have reported a direct association between PICALM rs3851179 and cognitive performance. The A allele, however, has been found to have a protective role as it participates in amyloid β clearance, whereas the GG genotype has been linked to poor cognitive performance and functional changes in the right inferior frontal gyrus and both superior frontal gyri; these regions are associated with episodic memory and executive function.3 In fact, carriers of the GG genotype have shown poorer performance in verbal fluency tasks.33 Although our frequency analysis showed an association between the CC genotype and poor performance in working memory tasks, this was not observed in non-parametric tests.
Our study found significant differences in short-term visual memory tasks between individuals with different CLU rs11136000 genotypes. Other studies have reported an association between CLU and poor episodic memory, particularly in the presence of certain APOE isoforms.31 Our study found no association between APOE and cognitive performance. This may be explained by the fact that the risk allele was not frequent in our sample (with the APOE 33 genotype being the most prevalent), which may have masked the true impact of other genotypes.
GWAS_14q rs11622883 is strongly correlated with AD due to its potential involvement in the pathogenesis of amyloid β deposition34; however, the association between this polymorphism and cognitive function is yet to be studied. Our frequency analysis hints at an association with performance in attention and information processing speed tasks; however, our results are merely exploratory and require confirmation by studies with larger samples. We also observed an association between the CC genotype of SORL1 rs11218304 and poor performance in planning, memory, and information processing speed tasks, although the association was not significant in non-parametric tests. This polymorphism has not previously been studied in the context of cognitive function, although its role in the pathophysiology of neurodegenerative disease makes it an interesting line of research.
We also observed an association between PVRL2 rs6859 and poor executive function, attention, and memory. PVRL2 has been associated with longevity,35 lower risk of AD, and slower disease progression.36 The APOC3-APOE-TOMM40-PVRL2 gene cluster, located on chromosome 19q13.32, may explain part of the variance in MoCA scores.37 However, the role of PVRL2 in this cluster requires further study.
Lastly, although TOMM40 is a significant candidate gene for AD and has been associated with cognitive performance38,39 and age at disease onset,40 our study found no significant associations between TOMM40 variants and cognitive function. Given its association with age of onset in our population,40 future studies should analyse the association between TOMM40 and clinical variables, including cognitive function.
Our study has several limitations. Firstly, although our exclusion criteria aimed to minimise the influence of variables associated with non-neurodegenerative cognitive impairment, data analysis did not control for such confounding variables as dyslipidaemia, arterial hypertension, diabetes mellitus, smoking, and sedentary lifestyle, which constitute important risk factors for neurodegenerative disease.41
Furthermore, although our results point to an association between cognitive performance and allele combinations of some polymorphisms, confidence intervals were wide, perhaps due to the small size of our sample, preventing us from extrapolating our results to the general population. Therefore, our results are exploratory and should be confirmed by larger studies into the influence of genetic variables on MCI in our population.
FundingThis study was funded by the Colombian Administrative Department of Science, Technology, and Innovation (2015).
Conflicts of interestNone.
We wish to thank all the participants and their families.
Please cite this article as: Cruz-Sanabria F, Bonilla-Vargas K, Estrada K, Mancera O, Vega E, Guerrero E, et al. Análisis de desempeños cognitivos y polimorfismos en SORL, PVRL2, CR1, TOMM40, APOE, PICALM, GWAS_14q, CLU y BIN1 en pacientes con trastorno neurocognitivo leve y en sujetos cognitivamente sanos. Neurología. 2021;36:681–691.