Glioma presents high incidence and poor prognosis, and therefore more effective treatments are needed. Studies have confirmed that long non-coding RNAs (lncRNAs) basically regulate various human diseases including glioma. It has been theorized that HAS2-AS1 serves as an lncRNA to exert an oncogenic role in varying cancers. This study aimed to assess the value of lncRNA HAS2-AS1 as a diagnostic and prognostic marker for glioma.
MethodsThe miRNA expression data and clinical data of glioma were downloaded from the TCGA database for differential analysis and survival analysis. In addition, pathological specimens and specimens of adjacent normal tissue from 80 patients with glioma were used to observe the expression of HAS2-AS1. The receiver operating characteristic (ROC) curve was used to analyze the diagnostic ability and prognostic value of HAS2-AS1 in glioma. Meanwhile, a Kaplan–Meier survival curve was plotted to evaluate the survival of glioma patients with different HAS2-AS1 expression levels.
ResultsHAS2-AS1 was significantly upregulated in glioma tissues compared with normal tissue. The survival curves showed that overexpression of HAS2-AS1 was associated with poor overall survival (OS) and progression-free survival (PFS). Several clinicopathological factors of glioma patients, including tumor size and WHO grade, were significantly correlated with HAS2-AS1 expression in tissues. The ROC curve showed an area under the curve (AUC) value of 0.863, indicating that HAS2-AS1 had good diagnostic value. The ROC curve for the predicted OS showed an AUC of 0.906, while the ROC curve for predicted PFS showed an AUC of 0.88. Both suggested that overexpression of HAS2-AS1 was associated with poor prognosis.
ConclusionsNormal tissues could be clearly distinguished from glioma tissues based on HAS2-AS1 expression. Moreover, overexpression of HAS2-AS1 indicated poor prognosis in glioma patients. Therefore, HAS2-AS1 could be used as a diagnostic and prognostic marker for glioma.
Los gliomas presentan una alta incidencia y un mal pronóstico, por lo que es necesario un tratamiento más efectivo. Algunos estudios han confirmado que los ARN no codificantes de cadena larga (ARNncl) regulan diferentes enfermedades, entre las que se incluyen los gliomas. Se ha postulado que HAS2-AS1 actúa como un ARNncl, con un efecto oncogénico en diferentes tipos de cáncer. Este estudio tiene como objetivo analizar el valor del ARNncl HAS2-AS1 como marcador diagnóstico y pronóstico de glioma.
MétodosDescargamos los datos clínicos y de expresión de micro-ARN de la base de datos del Atlas del Genoma del Cáncer (TCGA) para realizar el análisis diferencial y de supervivencia. También analizamos la expresión de HAS2-AS1 en muestras patológicas y muestras de tejido adyacente normal de 80 pacientes con glioma. Para analizar la capacidad diagnóstica y el valor pronóstico de HAS2-AS1 en el glioma, recurrimos a la curva ROC. También utilizamos curvas de Kaplan-Meier para evaluar la supervivencia de los pacientes con glioma con diferentes niveles de expresión de HAS2-AS1.
ResultadosLa expresión de HAS2-AS1 era significativamente mayor en las muestras patológicas que en el tejido normal. Las curvas de supervivencia demostraron que la sobreexpresión de HAS2-AS1 estaba relacionada con una menor supervivencia general y supervivencia libre de progresión. Algunos factores clínico-patológicos de los pacientes con glioma, como el tamaño del tumor y su grado, según la clasificación de la OMS, mostraron una correlación significativa con la expresión de HAS2-AS1 en los tejidos afectados. La curva ROC mostró un área bajo la curva de 0,863, lo que indica que la expresión de HAS2-AS1 posee un importante valor diagnóstico. El área bajo la curva de la supervivencia general estimada fue de 0,906; para la supervivencia libre de progresión estimada, de 0,88. Ambos valores muestran que la sobreexpresión de HAS2-AS1 se asocia con un mal pronóstico.
ConclusiónLos tejidos normales pueden distinguirse claramente de los tejidos afectados por glioma en función de la expresión de HAS2-AS1. Además, la sobreexpresión de HAS2-HS1 fue indicativa de mal pronóstico en los pacientes con glioma. Por tanto, HAS2-AS1 podría utilizarse como marcador diagnóstico y pronóstico en el manejo del glioma.
Glioma is one of the most common primary tumors in the central nervous system, accounting for about 40–50% of primary intracranial tumors. Glioma includes a variety of histologic features and the world health organization (WHO) has classified glioma using phenotypic and genotypic parameters since 2016 to make a more objective diagnosis of glioma.1 Furthermore, the tumor is graded from the lowest to the highest malignancy degree (I, II, III, and IV) according to its expected biological behavior. Grade I is characterized by benign cytology, grade II is characterized by medium cellularity without dysplasia or mitotic activity, and grade III is characterized by cellularity with hypoplasia and mitosis, grade IV is the same as grade III with additional proliferation and necrosis of microvessels.2 Besides grade I tumor can be cured by surgery, the rest of the glioma is hard to be cured by the existing treatment, and most patients experience relapse. Generally, patients with early diagnosis have a better prognosis, with a 5-year survival rate of 42–92%.3–5
It is found that one of the main reasons for the unsatisfactory survival of glioma after treatment is that currently, the detection indexes for glioma are generally still on the patient's age and tumor histology, leading to huge challenges in clinical prognosis, treatment identification and early diagnosis of patients.6 Therefore, a reliable biomarker for diagnosis and prognosis of glioma is urgently needed in clinic. Long non-coding RNAs (lncRNAs) are a type of RNAs whose transcript length is greater than 200 nucleotides. Studies have reported that lncRNAs can regulate gene expression at epigenetic, transcriptional and post-transcriptional levels through chromosome modification, transcriptional activation or interference.7 Some studies have indicated that lncRNAs are closely related to tumors, and there are significant differences in the expression of some lncRNAs in normal tissues and tumor tissues. Abnormally expressed lncRNAs may play an important role in the occurrence of tumors, and lncRNAs may become new markers for tumor diagnosis and evaluation of prognosis as well as therapeutic effect.8
Hyaluronan (HA) is one of the commonest glycosaminoglycan (GAG) in extracellular matrix (ECM), wherein hyaluronan synthase 2 (HAS2) is the main HAS participating in HA synthesis.9 Recent reports pointed out that HAS2 antisense 1 (HAS2-AS1) is a kind of lncRNA belonging to natural antisense transcript, and controls HAS2 via epigenetics.10 For instance, a study11 manifested that Sirtuin 1 represses nuclear translocation of NF-κB and lncRNA HAS2-AS1 expression to decrease HAS2 expression. HAS2-AS1 as a natural antisense RNA can generate heterodimer of HAS2 mRNA/HAS2-AS1 to foster HAS2 expression.12 Besides, HAS2-AS1 as a lncRNA is proved to impose an oncogenic role in multiple cancers including glioblastoma,13 oral squamous cell carcinoma,14 and epithelial ovarian cancer.15 A study16 authenticated that transcription factor USF1 binds to lncRNA HAS2-AS1 promotor, thereby facilitating invasive and migratory capabilities of glioma. The other study also exhibited that transcription factor C/EBPβ stimulates human fetal lung fibroblast-1 (HFL-1) cell migration, proliferation, and inflammation through activating lncRNA HAS2-AS1 in hypoxia.17 Nonetheless, the specific role of lncRNA HAS2-AS1 in glioma is not clear.
Here, we analyzed the expression level of HAS2-AS1 in 80 clinical samples, constructed receiver operating characteristic (ROC) curves to evaluate the diagnostic and prognostic value of HAS2-AS1 in glioma, and provided a new direction for the clinical treatment of glioma.
MethodsBioinformatics analysisGene Expression Quantification data and miRNA Expression Quantification data of TCGA_LGG and TCGA_GBM were downloaded from TCGA database (https://portal.gdc.cancer.gov/), and glioma tissues along with normal tissues of 670 patients were obtained. EdgeR was used for differential analysis with |logFC|>2 and padj<0.05 as the threshold to find out the differentially expressed lncRNAs (DElncRNAs), and survival analysis was carried out on DElncRNAs.
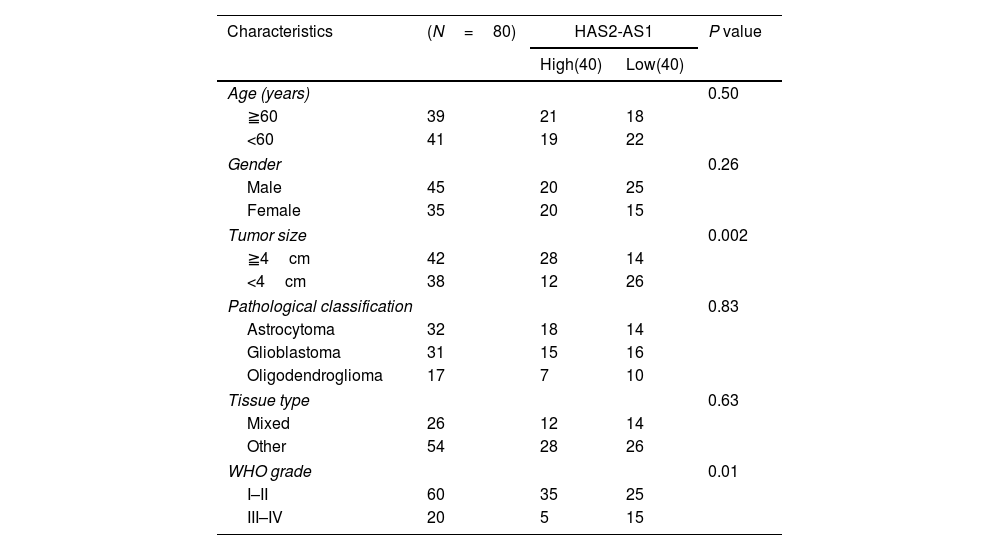
Sample selectionThis study collected tumor and adjacent normal tissue samples from 80 patients with glioma admitted to Tangshan Gongren Hospital from January 2015 to January 2018. All patients had not received any treatment before the operation. The tissue samples of the patients were obtained during the operation and all specimens were stored in liquid nitrogen for rapid freezing after resection. The tumor histology of each sample was assessed by senior pathologists. The specimens were obtained with the approval and consent of the ethics committee of Tangshan Gongren Hospital, and all patients had signed the informed consent. The patients were classified into the WHO (I–II) grade and the WHO (III–IV) grade according to the WHO classification standard. The pathological factors of the patients including gender, age, tumor size, pathological classification, tissue type and WHO grade were listed in Table 1.
Clinicopathologic characteristics of the glioma patient.
| Characteristics | (N=80) | HAS2-AS1 | P value | |
|---|---|---|---|---|
| High(40) | Low(40) | |||
| Age (years) | 0.50 | |||
| ≧60 | 39 | 21 | 18 | |
| <60 | 41 | 19 | 22 | |
| Gender | 0.26 | |||
| Male | 45 | 20 | 25 | |
| Female | 35 | 20 | 15 | |
| Tumor size | 0.002 | |||
| ≧4cm | 42 | 28 | 14 | |
| <4cm | 38 | 12 | 26 | |
| Pathological classification | 0.83 | |||
| Astrocytoma | 32 | 18 | 14 | |
| Glioblastoma | 31 | 15 | 16 | |
| Oligodendroglioma | 17 | 7 | 10 | |
| Tissue type | 0.63 | |||
| Mixed | 26 | 12 | 14 | |
| Other | 54 | 28 | 26 | |
| WHO grade | 0.01 | |||
| I–II | 60 | 35 | 25 | |
| III–IV | 20 | 5 | 15 | |
Two μg of total RNA was isolated from tissue samples or cultured cells using Trizol reagent (Invitrogen, Carlsbad, California) following manufacturer's instructions. The isolated RNA was treated using PrimeScript RT Master Mix (Takara, Dalian, China) to prepare complementary DNA (cDNA) for PCR analysis. PCR reaction was performed on LightCycler 480 system using TB GreenPremix Ex Taq II (Takara). The corresponding primers were as follows: HAS2-AS1: 5′-GACAGGACCTTGAAGACTGGG-3′ (forward) and 5′-GGGATGGAGGTCAGCAACAA-3′ (reverse); GAPDH: 5′-ACAACTTTGGTATCGTGGAAGG-3′ (forward) and 5′-GCCATCACGCCACA GTTTC-3′ (reverse). GAPDH was the internal reference of HAS2-AS1. HAS2-AS1 expression level was calculated by 2−ΔΔCt method.
Statistical analysisGraphPad Prism (version 7.0, GraphPad software, Inc., La Jolla, CA, USA) and SPSS software (version 23.0, SPSS Inc., Chicago, IL) were used for data analysis. Kaplan–Meier curve was used to analyze the overall survival (OS) of glioma patients. Chi-square test was used to examine the correlation between HAS2-AS1 expression level and clinicopathological parameters. P<0.05 indicated a statistically significant difference. The diagnostic ability of HAS2-AS1 in glioma was detected by ROC curve. ROC curve was drawn by using SPSS 23.0, and the area under the curve (AUC) was calculated using rank-sum test. The larger the area, the higher the judgment value. The “Youden index” was determined using the best cutoff value. The maximum value of the index was the best boundary value.
Follow-up visit80 patients were followed up at least once. The follow-up was mainly to detect the expression of HAS2-AS1 in the patients and observe the prognostic effect. The follow-up period began from the first day of treatment to May 31, 2018. A total of 80 patients were followed up for 6–36 months and the median follow-up time was 12.3 months. One patient was lost to follow-up, with a total follow-up rate of 98.75%. OS refers to the duration time from the first day after surgery to death or from the last follow-up time to the time of loss of follow-up. Progression-free survival (PFS) is the period from the first day of surgery to recurrence and metastasis of patients.
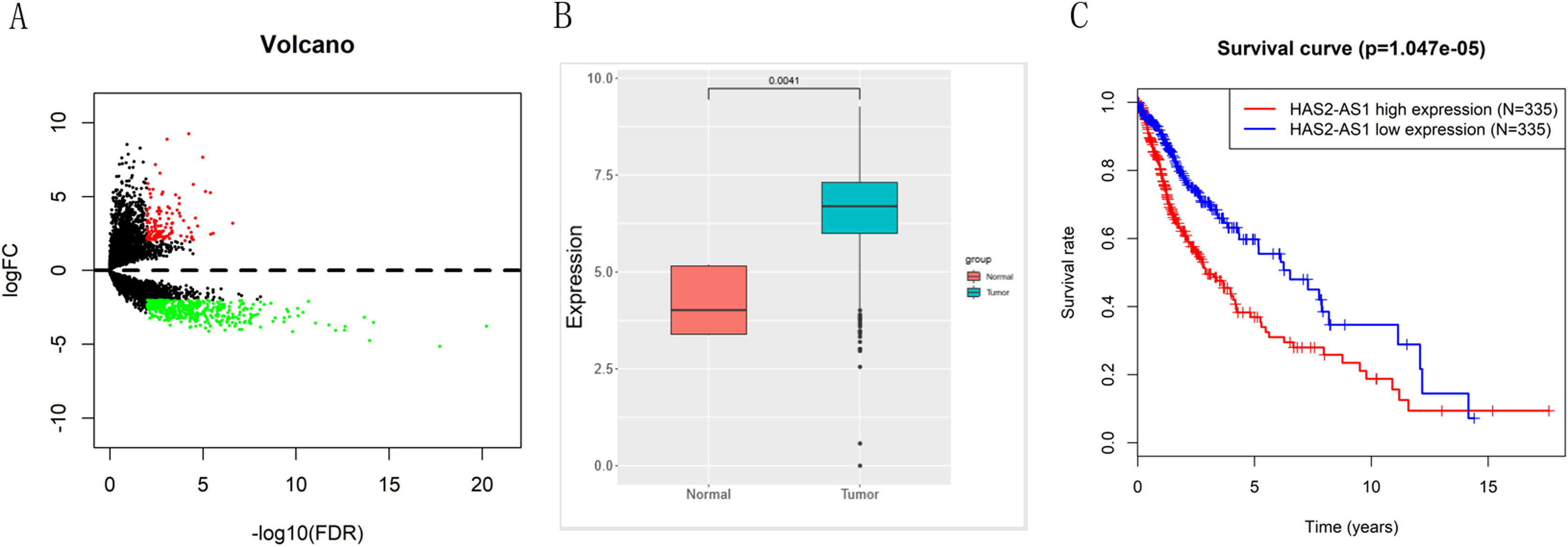
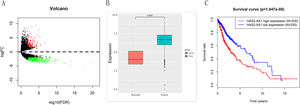
ResultsResults of bioinformatics analysisEdgeR differential analysis results showed that a total of 557 DElncRNAs were obtained (Fig. 1A), among which HAS2-AS1 was highly expressed in glioma (Fig. 1B). Survival analysis indicated that high expression of HAS2-AS1 indicated an extremely high survival risk, suggesting that HAS2-AS1 was of great survival significance (Fig. 1C). Hence, HAS2-AS1 was likely to be a potential diagnostic and prognostic marker for glioma.
TCGA expression profile analysis reveals that lncRNA HAS2-AS1 is significantly overexpressed in glioma and of great survival significance. (A) The volcano map of edgeR differential analysis results; (B) HAS2-AS1 expression level in glioma and normal tissues in TCGA; (C) Survival rate of patients with high or low HAS2-AS1 expression were compared.
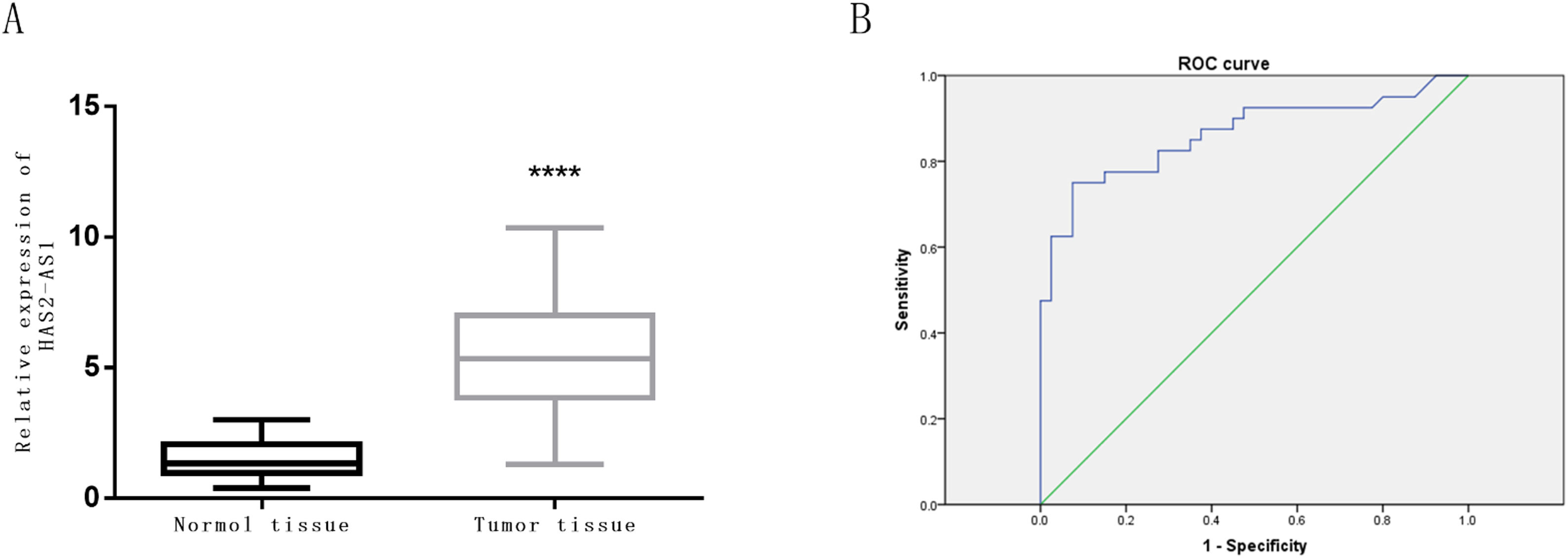
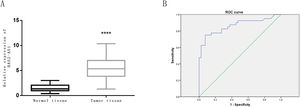
The expression level of HAS2-AS1 in clinical glioma tissue and normal tissue of 80 patients was evaluated by qRT-PCR. The results revealed that the relative mRNA expression of HAS2-AS1 in glioma tissues was significantly higher than that in normal tissues (P<0.0001, Fig. 2A). ROC curves were plotted based on HAS2-AS1 expression in glioma and normal tissues in the study. It could be concluded from the ROC curves that AUC value was 0.863 with the cut-off value of 5.3, sensitivity of 75%, specificity of 92.5% and Youden index of 67.5%. HAS2-AS1 was confirmed to possess good sensitivity and specificity in the diagnosis of glioma, along with good diagnostic value indicated by the AUC and Youden index. Furthermore, glioma tissues could be distinguished from normal tissues according to the expression level of HAS2-AS1 (Fig. 2B).
Expression level of HAS2-AS1 in normal tissues and glioma tissues and its diagnostic value. (A) The expression level of HAS2-AS1 in clinical normal tissues (n=80) and glioma tissues (n=80); (B) The ROC curves were plotted for evaluation of the diagnostic value of HAS2-AS1 expression in glioma (the sensitivity is 75% and the specificity is 92.5%). AUC refers to area under the curve. **** P<0.0001.
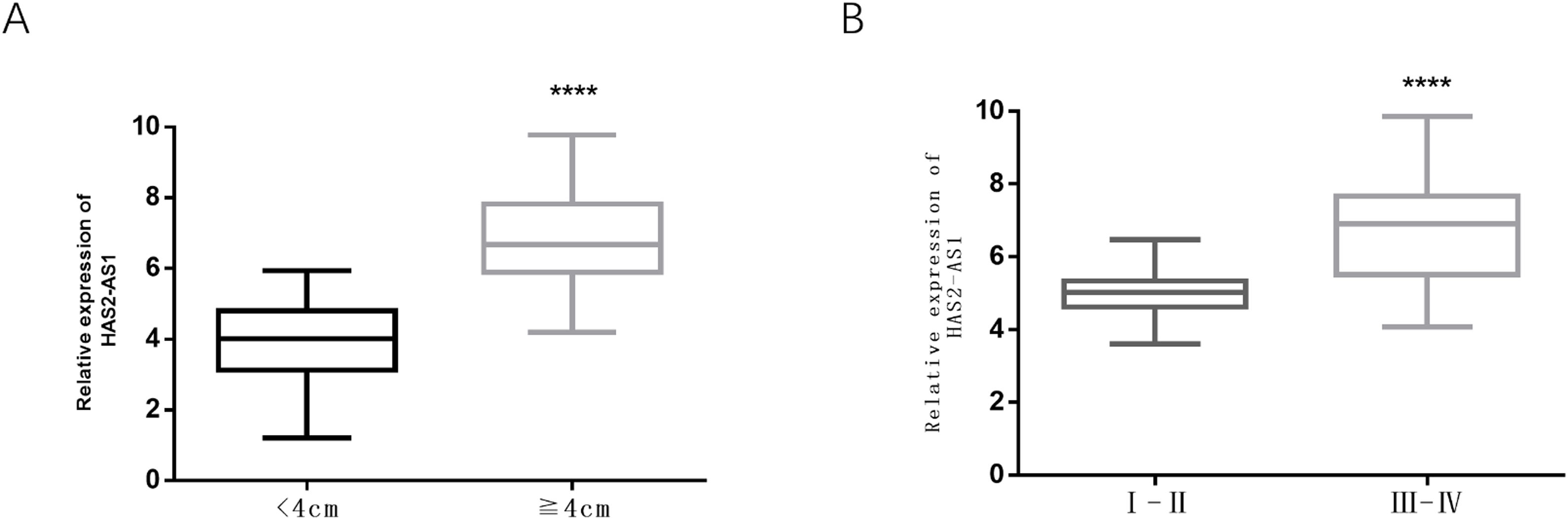
Here, glioma patients were divided into high-expression group (n=40) and low-expression group (n=40) according to median HAS2-AS1 expression level, and the correlation between HAS2-AS1 expression level and clinicopathological factors in glioma patients was detected by chi-square test. There was no significant correlation between HAS2-AS1 expression level and age, gender, pathological classification, and tissue type (P>0.05). While tumor size (P=0.002) and WHO grade (P=0.01) were remarkably correlated with HAS2-AS1 expression (Table 1).
Tumor size and WHO grade are significantly correlated with HAS2-AS1 expression in gliomaIt had been concluded from Table 1 that tumor size and WHO grade were dramatically correlated with HAS2-AS1 expression level. Box plots were used to demonstrate the specific correlation. It could be observed that the expression level of HAS2-AS1 in glioma tissues with a tumor size of more than 4cm was obviously higher than that in glioma tissues with a size of less than 4cm (Fig. 3A). Additionally, the expression of HAS2-AS1 in WHO grade III–IV glioma tissues was also significantly higher than that in the I–II glioma tissues (Fig. 3B).
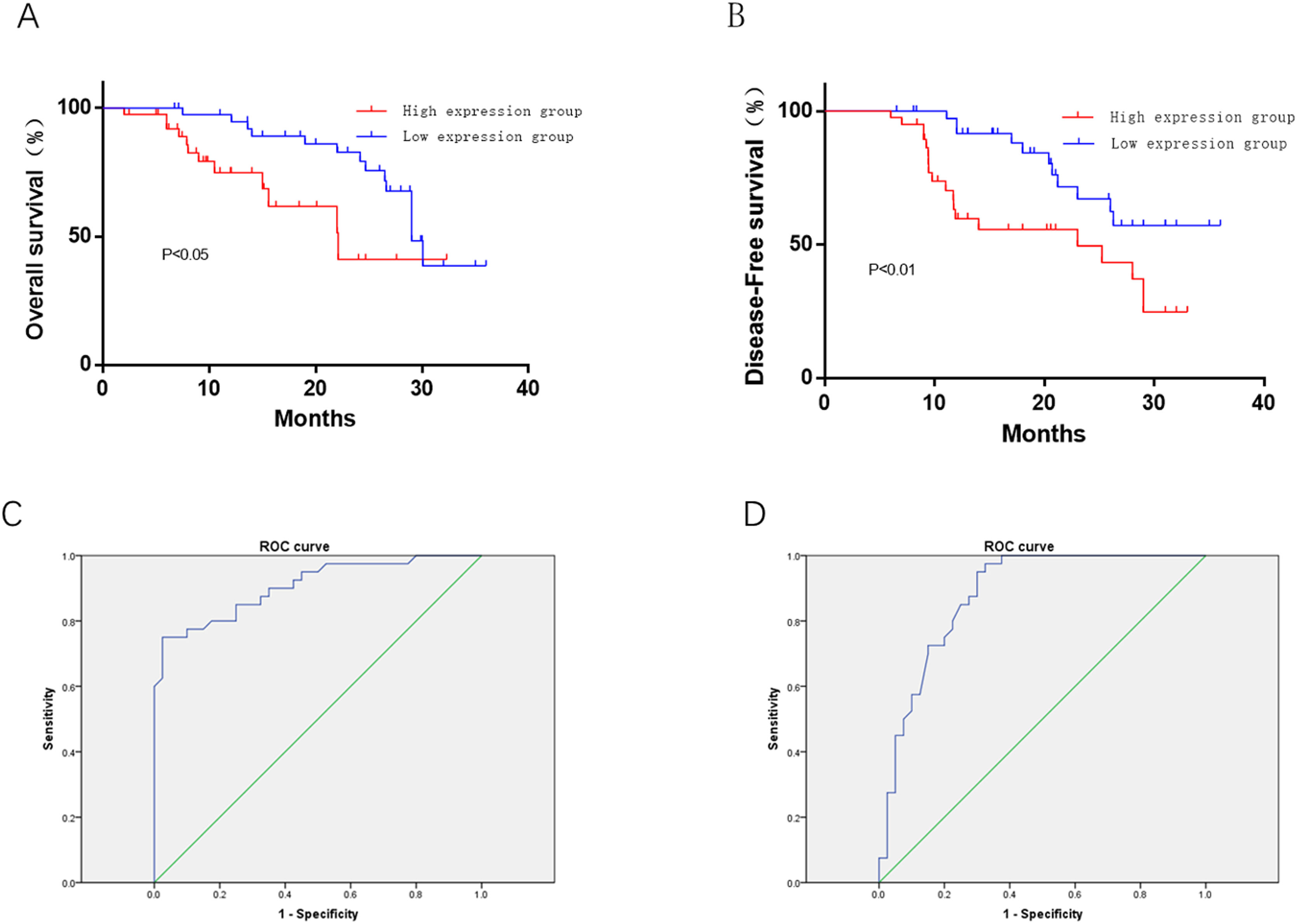
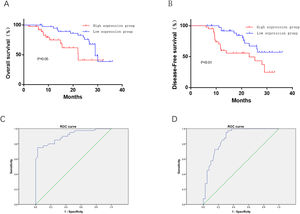
High expression of HAS2-AS1 indicates poor prognosis and poor survival rateSurvival analysis revealed that patients with low HAS2-AS1 expression share an relatively optimal survival (P<0.01), suggesting that high HAS2-AS1 expression was related to poor OS (Fig. 4A). PFS curves exhibited that patients with low HAS2-AS1 expression had significantly higher PFS, revealing that high expression of HAS2-AS1 suggested poor PFS (Fig. 4B). ROC analysis for the prognostic value of HAS2-AS1 in OS of glioma patients showed the AUC value of 0.906, cut-off value of 5.3, sensitivity of 75%, specificity of 97.5%, and Youden index of 72.5% (Fig. 4C). ROC analysis for the prognostic value of HAS2-AS1 in PFS of glioma patients showed the AUC value of 0.88, cut-off value of 4.9, sensitivity of 95%, specificity of 85%, and Youden index of 65.5% (Fig. 4D). The above data suggested that high expression of HAS2-AS1 was associated with poor prognosis in glioma patients and had good prognostic value.
Effect of HAS2-AS1 expression on OS and PFS in glioma patients and corresponding prognostic value. The (A) OS and (B) PFS of patients with high HAS2-AS1 expression were compared with those of patients with low expression; the predictive value of HAS2-AS1 expression in (C) OS (sensitivity of 75%, specificity of 97.5%) and (D) PFS (sensitivity of 95%, specificity of 85%) of glioma patients.
Glioma is a disease in the central nervous system with the highest mortality and is the leading cause of brain diseases related deaths in children and adults.18 Its incidence increases with age and peaks between 75 and 84.19 Despite that most aggressive treatment methods have been applied for glioma, the survival and prognosis remain poor, with a median survival of 12–15 months and a low 5-year survival rate.20,21 The traditional treatment methods for glioma include surgical treatment, radiotherapy and chemotherapy, etc. However, due to the invasive growth of glioma, the boundary of tumor tissues and adjacent normal brain tissues is unclear, so it is difficult to completely resect the glioma tumor, resulting in poor treatment and common recurrence. Radiotherapy and chemotherapy are often used as adjuvant means after the surgery, but they could not specifically kill tumor cells, and may also produce toxic and side effects on the central nervous system, so the treatment of glioma is one of the difficulties in neurosurgical research.22–24 In order to change the clinical status that diagnosis and prognosis prediction of glioma disease can only depend on patient's age and tumor histological phenotypes, a reliable biomarker is in need for clinical diagnosis of glioma and the prognosis prediction after surgical treatment. LncRNAs can be applied in glioma and it has been reported that LINC00599 is down-regulated in glioma tissues and cell lines, and is related to WHO grade and prognosis of glioma patients.25 Increased expression of lncRNA AB073614 may be a biomarker of poor prognosis in glioma.26 LncRNA MEG3 plays an important role in glioma cell proliferation, apoptosis and autophagy, and this gene is also significantly associated with the prognosis of glioma patients.27 All the above studies have proved that 1ncRNAs play a vital role in glioma.
Abnormal expression of the human hyaluronan synthase 2 (HAS2) gene has been linked to malignant tumors, pulmonary arterial hypertension, osteoarthritis, asthma, thyroid dysfunction, and large organ fibrosis.28 Studies have indicated that HAS2-AS1 knockout can functionally inhibit the proliferation, invasion and tumor growth of ovarian cancer cells in vitro and in vivo, while overexpression of HAS2-AS1 indicates poor clinical prognosis of patients with ovarian cancer.15 It is found that the reduction of HAS2-AS1 inhibits the migration and invasion of glioma cells in vivo and in vitro.13 In addition, many investigations reported that HAS2-AS1 is involved in the regulation mechanism of ceRNA network to modulate the proliferation and migration of tumor cells.29,30
Glioma tissues and adjacent normal tissues from 80 patients were involved in the study. The results exhibited that the expression of HAS2-AS1 in glioma tissues was significantly higher than that in normal tissues, which was consistent with the bioinformatics analysis results of HAS2-AS1 and glioma.30 Moreover, the tumor size and WHO grade of glioma were correlated with the expression of HAS2-AS1 in tumors, which was consistent with the results of the review and meta-analysis of lncRNAs and glioma.31 The OS of patients with high HAS2-AS1 expression was remarkably lower than that of patients with low expression. The ROC curves proved that HAS2-AS1 had better diagnostic and prognostic value in glioma. The survival curves suggested that high expression of HAS2-AS1 was associated with poor survival. However, only 80 patients were included in this study, and the sample size was small, leading to the inaccuracy of HAS2-AS1 expression in glioma. In addition, isocitrate dehydrogenase (IDH) 1/2 mutation is the golden standard for glioma diagnosis. Nevertheless, this study was limited by the absence of discussion about the relationship between HAS2-AS1 and IDH1/2. In future studies, the sample size should be enlarged to determine a more accurate value for the diagnosis and prognosis of glioma.
In summary, HAS2-HA1 can be used as a clinical biomarker for the diagnosis and prognosis prediction of glioma.
Authors’ contributionsAW Y and X G mainly contributed to revising the article. JS G, XH G and AW Y contributed to data analysis. J L, YY Z and GM R contributed to drafting and modifying the article. JT W and K Z contributed to final approval of the version to be published. DC W agreed to be accountable for all aspects of the work.
Ethics approval and consent to participateThis study was conducted in accordance with the Helsinki Declaration II and was approved by the Research Ethics Committee of Tangshan Gongren Hospital. Written informed consent was obtained from individual or guardian participants. (GRYY-LL-2019-66).
Consent for publicationAll authors consent to submit the manuscript for publication.
Availability of data and materialsThe data used to support the findings of this study are included within the article. The data and materials in the current study are available from the corresponding author on reasonable request.
FundingNot applicable.
Conflict of interestThe authors declare no conflicts of interest.